Hematopoietic stem/progenitor cell transplantation is the main treatment option for hematological malignancies and disorders. One strategy to solve the problem of low stem cell doses used in transplantation is pre-transplant expansion. We hypothesized that using fibronectin-coated microfluidic channels would expand HSPCs and keep self-renewal potential in a three-dimensional environment, compared to the conventional method. We also compared stem cell homing factors expression in microfluidic to conventional cultures.
Materials and methodsA microfluidic device was created and characterized by scanning electron microscopy. The CD133+ cells were collected from cord blood and purified. They were subsequently cultured in 24-well plates and microfluidic bioreactor systems using the StemSpan serum-free medium. Eventually, we analyzed cell surface expression levels of the CXCR4 molecule and CXCR4 mRNA expression in CD133+ cells cultured in different systems.
ResultsThe expansion results showed significant improvement in CD133+ cell expansion in the microfluidic system than the conventional method. The median expression of the CXCR4 in the expanded cell was lower in the conventional system than in the microfluidic system. The CXCR4 gene expression up-regulated in the microfluidic system.
ConclusionUtilizing microfluidic systems to expand desired cells effectively is the next step in cell culture. Comparative gene expression profiling provides a glimpse of the effects of culture microenvironments on the genetic program of HSCs grown in different systems.
Over the years, hematopoietic stem/progenitor cell (HSPC) transplantation has treated a variety of hematological malignancies and non-hematological disorders, as well as immunodeficiency syndromes, inborn errors of metabolism and autoimmune diseases. Umbilical cord blood (UCB) is an excellent source of HSPCs with a lower rate of graft versus host disease (GVHD).1 Unfortunately, hematopoietic stem cells (HSCs) isolated from single cord blood are low in number and they need more time to lodge in the bone marrow to start producing blood cells than peripheral blood or bone marrow stem cells.2 For this reason, to date, mostly children have been able to reap the benefits of the UCB-HSC transplantation, while adults are restricted in using this method.3
One strategy to overcome the low cell dose and to improve the homing period is to develop an efficient method for in vitro UCB-HSC expansion prior to transplantation.4,5 Most of the present hematopoietic culture conditions are conventional strategies that use static, two-dimensional (2D) culture devices, such as wells and T-flasks. There are specialized structures in the bone marrow called niches. Niches, ample with growth factors and an extracellular matrix (ECM), are responsible for preserving stem cells and their ability for self-renewal. It has been demonstrated that three-dimensional (3D) cultures, which are more comparable to native HSC niches, expand cells more efficiently than traditional 2D cultures. In 2D cultures, cell-cell and cell-environment interactions are not established and the spatial importance of the ECM in cell fate is ignored.5,6 Also, the lack of mixing in the static culture results in critical concentrations of pH, nutrients, oxygen and cytokines, hindering optimal cell expansion. This calls for the invention of new methods that can not only tightly control every aspect of the cell culture, but also mimic the environmental conditions cells feel more comfortable growing in. Microfluidic bioreactors, which provide an accurate spatio-temporal condition close to the cellular environment, help scientists in this matter. With a continuous flow that provides a fresh medium and removes metabolic waste and smaller dimensions that reduce costs and make for an easier management, these bioreactors achieve these goals.7,8 A downfall in the traditional expansion of CD133+ cells is that besides stimulating proliferation, it drives stem cells to differentiate into a more mature form. Thus, maintaining a higher proportion of HSPCs when the total cell number expands is an important factor in ex vivo expansion. As explained, ECM is an integral part of cell expansion and fibronectin (FN) is one of its important components. Specific domains of fibronectin mediate adhesion and migration of early murine erythroid progenitors. They also, to some extent, convey the adhesion of progenitors to the stroma.9 To test the effect of fibronectin in this study, we expanded UCB-extracted CD133+ cells in an environment with the aforementioned conditions.
Migration of HSCs through the bloodstream toward bone marrow (BM) niches requires active navigation called the homing mechanism. A better understanding of the homing mechanism nullifies the need for a huge number of cells for transplantation, which in turn translates to better results.10,11 Undoubtedly, the CXCR4/SDF-1 signaling axis is important and cytokine-expanded cultures are believed to alter homing receptors, such as the CXCR4.9,10,12,13
In the present study, we used microfluidic channels that not only imitate the three-dimensional structure of niches, but are also thought to increase the homing receptor of the CXCR4.14 An ideal microenvironment was fabricated and efficient expansion of HSCs will be performed to reach a sufficient number applicable for cell therapy and transplantation.
In this study, we evaluated the role of fibronectin, an essential molecule in cell proliferation, by expanding cord blood stem cells in microfluidic covered with fibronectin. We also investigated the effect 3D and conventional 2D expansion conditions have on the UCB-HSC expansion and differentiation. Additionally, CXCR4 levels were evaluated in each of these conditions.
Materials and methodsMicrofluidic fabricationTo build microcavities, we used the soft lithography technique. Cost-efficiency and small detail production are some advantages this method offers. The microfluidic pattern was created and printed in high resolution. The pattern consisted of microchannels and microcavities. The generated transparency mask was used in the photolithography of the SU-8 photoresist (Microchem, USA). A layer of negative photoresist was spin-coated onto the 100-mm glass wafer (Shin-Etsu Group, Japan). The master was developed by immersion of the SU-8 wafer into developer propylene glycol methyl ether acetate (PGMEA) (Sigma, USA) under agitation for 10 min to remove the non-cross-linked photoresist. The polydimethylsiloxane (PDMS) (15:1 elastomer: curing agent; Sylgard 184 Silicone elastomer kit; Dow Corning, USA) was poured over the master and incubated overnight in an oven at 60o C. The PDMS replica was oxidized by plasma treatment to make the PDMS mold surface hydrophilic. The negatively charged PDMS surface was exposed to an ethylene dichloride/N-hydroxysuccinimide (EDC/NHS) solution (10 mg/ml) for 24 h and then rinsed thoroughly with distilled water. Diluted fibronectin solution was added to the PDMS surface and incubated for 24 h at 4 °C. The microcavities and microchannels system was brought into a cell culture cabinet and then sterilized by rinsing with 70% ethanol and rinsed thoroughly with sterile phosphate-buffered saline (PBS). Pneumatic micropumps were used to transport the cells and media in the microchannels.
Cord blood cell preparationHealthy child-bearing mothers gave informed consent, approved by the local Ethical Committee of the Tarbiat Modares University, for cord blood donation. Cord blood samples were collected at the time of birth. Briefly, after the baby had been delivered, the cord was clamped at both distal ends and cut. A needle was injected into the umbilical vein at a prepared site to harvest the UCB into a sterile 250 ml blood bag (BesaatQom Iran) containing 21 ml of anticoagulant citrate phosphate dextrose (CPD) solution by gravity flow. The cord blood bag was gently rotated to mix the anticoagulant with the blood. The collected UCB was immediately transferred to our laboratory. The UCB unit was processed within 2 h. Isolation of UCB-mononuclear cells (UCB-MNCs) was performed. Briefly, red blood cells were depleted by hydroxyethyl starch (Fresenius Kabi) within 1 h. Subsequently, the UCB-MNCs were isolated from the whole blood by density gradient separation medium (1.077 g/ml, Ficoll-Hypaque). In this procedure, red blood cells and plasma were removed and an MNC fraction containing hematopoietic stem cells was isolated. Positive selection of CD133 antigen-carrying cells was performed by the MidiMACS (MiltenyiBiotech, Germany) magnetic separation columns according to the protocols provided by the manufacturer. To further purify CD133+ cells, isolated cells were subjected to a second column separation. The purity of CD133+ cells in the selected population was above 95%. The CD133+ cell viability and number were determined by trypan blue in a hemacytometer.
Flow cytometryTo determine the purity of CD133 cells that were isolated with the magnetic-activated cell sorting (MACS) and cells expanded with different methods, cells were harvested and washed in a PBS containing 1% of Stain Buffer (FBS) and were each labeled with 10µl of phycoerythrin (PE)-conjugated anti-human CD133 (Miltenyi Biotech, Germany), following manufacturer instructions. Isotype controls were included to confirm specificity. After a wash, the Becton Dickinson FACSCalibur flow cytometer (Becton Dickinson, San Jose, CA, USA) was used to distinguish the fluorochrome antibody-tagged cells. Six thousand to 10,000 events were counted. The FACSCalibur data were analyzed using the Cyflogic (CyFlo Ltd, FINLAND).
Cell cultureThe CD133+ cells were seeded at 104 cells/ml in 24-well tissue culture plates by the conventional method and 104 cells/ml in the microfluidic bioreactor system in stem span serum-free medium (Stem Cell Technologies Inc., Vancouver, Canada) and were cultured for 12 days. Both mediums were supplemented with the proper mixture of growth factors and cytokines (50 ng/ml recombinant human stem cell factor, 20 ng/ml recombinant human thrombopoietin and 50 ng/ml recombinant human FLT3-L). All the cytokines were purchased from R&D Systems (Minneapolis, USA). The conventional culture was performed, as the control, in 24-well tissue culture plates (Nunc, Denmark) with 2 ml of culture medium per well. Both cultures were maintained at 37ºC in a humidified incubator with 5% CO2. The medium was replaced with the cytokine-containing fresh medium every 3 days. After culturing for 12 days, all expanded cells under conventional conditions were harvested and counted with a hemocytometer. In the microfluidic device, the covering layer of PDMS was removed aund the content of desired micro-cavities was harvested with a micropipette by flushing with sterile phosphate-buffered solution with ethylenediaminetetraacetic acid (PBS-EDTA).
Colony assaysFor the colony-forming cell (CFC) assay, 12 days after culture, cells were assayed for normal hematopoietic progenitor cell differentiation using an in vitro methylcellulose-based CFC assay (MethoCult® H4435 Enriched, StemCell Technologies) as per manufacturer instructions. Cells were seeded at 1 × 104 cells/ml in 24-well plates containing Iscove Modified Dulbecco Media (IMDM), 20% of FBS, 0.9% of methylcellulose and required cytokines. The plates were incubated at 37°C for 14 days in a humidified atmosphere containing 5% of CO2. After 14 days, colonies were counted using an inverted microscope. Colonies containing more than 50 cells were scored as colony-forming units (CFUs).
The long-term culture-initiating cell (LTC-IC) assay (Stem Cell Technologies) was performed according to manufacturer instructions. In brief, mitomycin C-treated feeder cells of murine mesenchymal stem cells were established in 35 mm tissue culture dishes at a seeding density of 3 × 105 cells/ml. Three hours after treatment with mitomycin C, the superficial layer was exchanged with a suspension of 5 × 105 cells/ml in the human long-term culture medium (HLTM) and incubated at 37°C, 5% of CO2 for 35 days. Once a week, half of the medium was extracted and replaced by new HLTM and fresh hydrocortisone.
Following 5 weeks of culture at 37°C, the contents of each dish were harvested, and 5×104 cells were plated into methylcellulose-containing dishes (MethoCult, H4230; StemCell Technologies). The dishes were cultured at 37°C, 5% of CO2, for 18 days until cells were scored.
RNA extraction and cDNA synthesisTotal cellular ribonucleic acid (RNA) of freshly isolated cells (day 0), cells expanded in 24-well tissue culture plates (day 12) and cells expanded in the microfluidic system (day 12) was extracted using the RNeasy Mini Kit (Qiagen, Valencia, CA) according to the protocols provided by the manufacturer. The quantity and purity of the RNA were determined by spectrophotometric analysis at 260 nm and 280 nm and the quality of all RNA samples was assessed by electrophoresis in 1.2% agarose gel. Subsequently, the complementary deoxyribonucleic acid (cDNA) was synthesized using the RevertAid™ First Strand cDNA Synthesis Kit (Fermentas, USA).
Reverse transcriptase-qPCRThe quantitative reverse transcription polymerase chain reaction (RT-qPCR) was performed using cDNA pools from freshly isolated and expanded CD133+ cells. RT-qPCR assays were developed to determine CXCR4 expression levels using the Sybr Green method. All RT-qPCR reactions were performed on the STEP ONE ABI platform (Applied Biosystems, USA). Gene-specific primers were designed according to Primer-Express software. Forward and reverse primers were designed in different exons or exon-exon junction to eliminate DNA contamination (Table 1). The Sybr Green Master Mix was purchased from ABI (SYBR green PCR kit; Applied Biosystems, Foster City, CA). The relative initial amount of mRNA of a particular gene was extrapolated from the standard curve. To plot the standard curve, we used a pool of all the samples serially diluted in four log2 steps and run in parallel to the samples. The total volume of each reaction was 10 μl, containing 300 nM of each forward and reverse primer and 125ng of cDNA. Appropriate negative controls were run for each reaction. The PCR amplification included the first step of 10 min at 95°C of denaturation, followed by 45 cycles of amplification at 95°C for 15 s and 60°C for 60 s. All of the reactions were performed in triplicate. The relative obtained amounts were normalized to the housekeeping gene beta-actin. The relative change in gene expression was calculated by dividing each gene expression signal by that of the internal standard (beta-actin) in each sample.
Scanning Electron Microscopy (SEM)Specimens were prepared for SEM analysis on day 4 using the following protocol. The superficial medium was gently removed and cell-microfluidic constructs were fixed two times. First, in the PBS with 2.5% of glutaraldehyde for 90 min at 4°C and then a second fixation in 1% osmium tetroxide for 2 h at 4 °C. Samples were dehydrated by soaking in a graded ethanol series of 30%, 50%, 70%, 90% and 100%. Samples were finally vacuum dried. The construct was mounted on an SEM specimen stub, sputter-coated with gold-palladium and imaged by SEM (Electronic Engineering College, Tehran University).
Statistical analysisThe percentage of CXCR4 positive cells was analyzed using the quadrant statistics of the dot plot (co-expression analysis) and expressed as the mean ± standard error in the mean of at least three experiments. Differences between populations of cells were analyzed using a two-tailed Student's t-test.
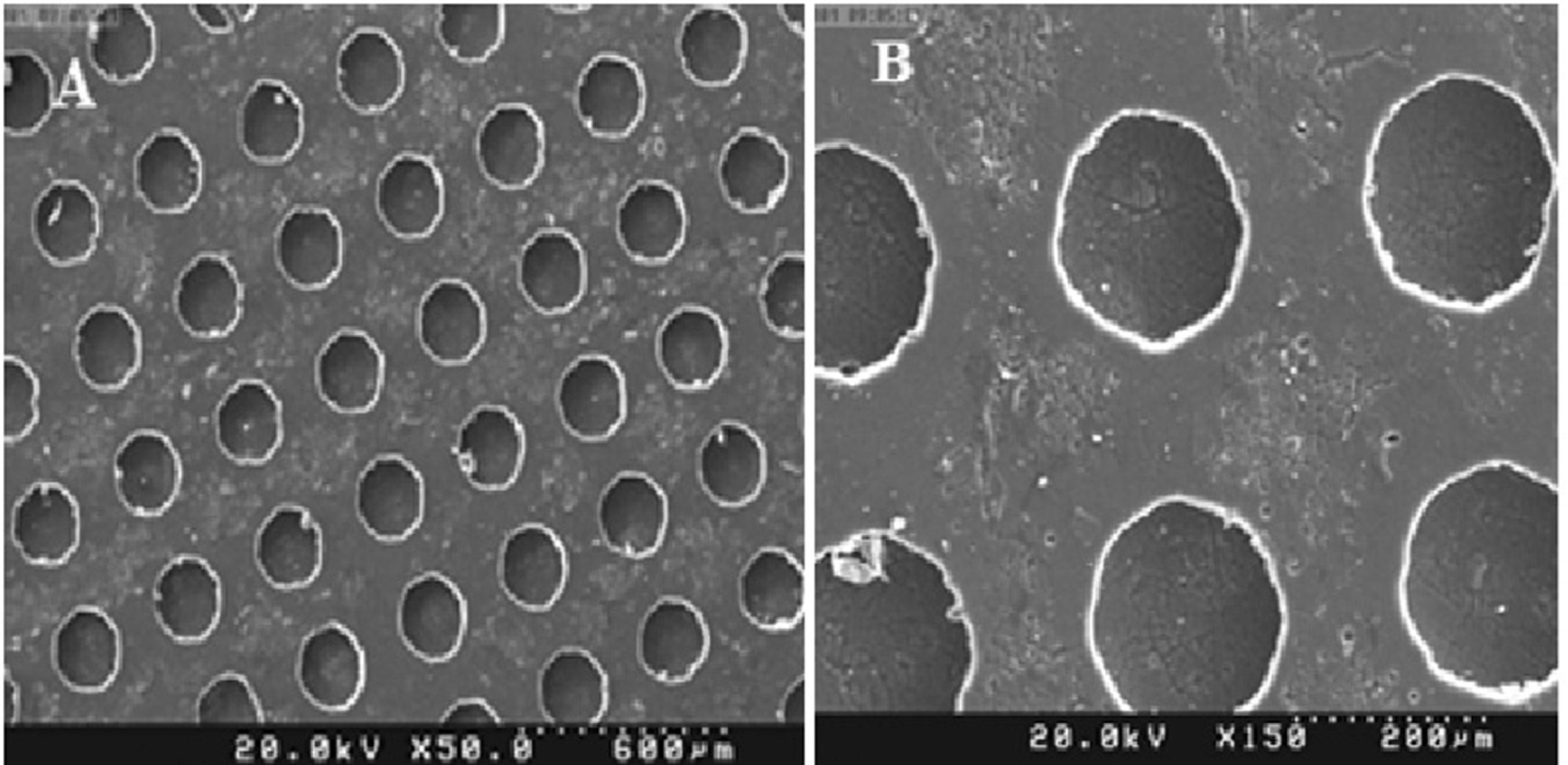
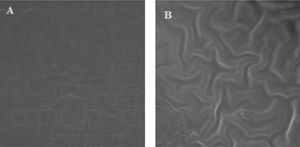
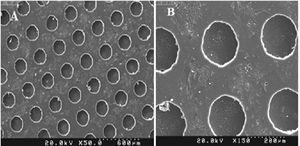
ResultsMicrofluidic device fabrication and surface characterizationFunctional FN-coated microcavities were fabricated and characterized for their usage in the investigation of UCB-HSC expansion. The microstructure (LPDMS) molds were replicated from lithographically designed silicon master structures. The human fibronectin fibrils were covalently attached via EDC/NHS onto the PDMS replicas containing channels and micro-cavities of approximately 150µm in diameter. The homogeneity of the FN layers was analyzed using scanning electron microscopy, showing a smooth surface on microcavities (Figure 1). Additionally, results from the SEM imaging showed the microfluidic bioreactor has separate, symmetrical cavity-like structures (Figure 2).
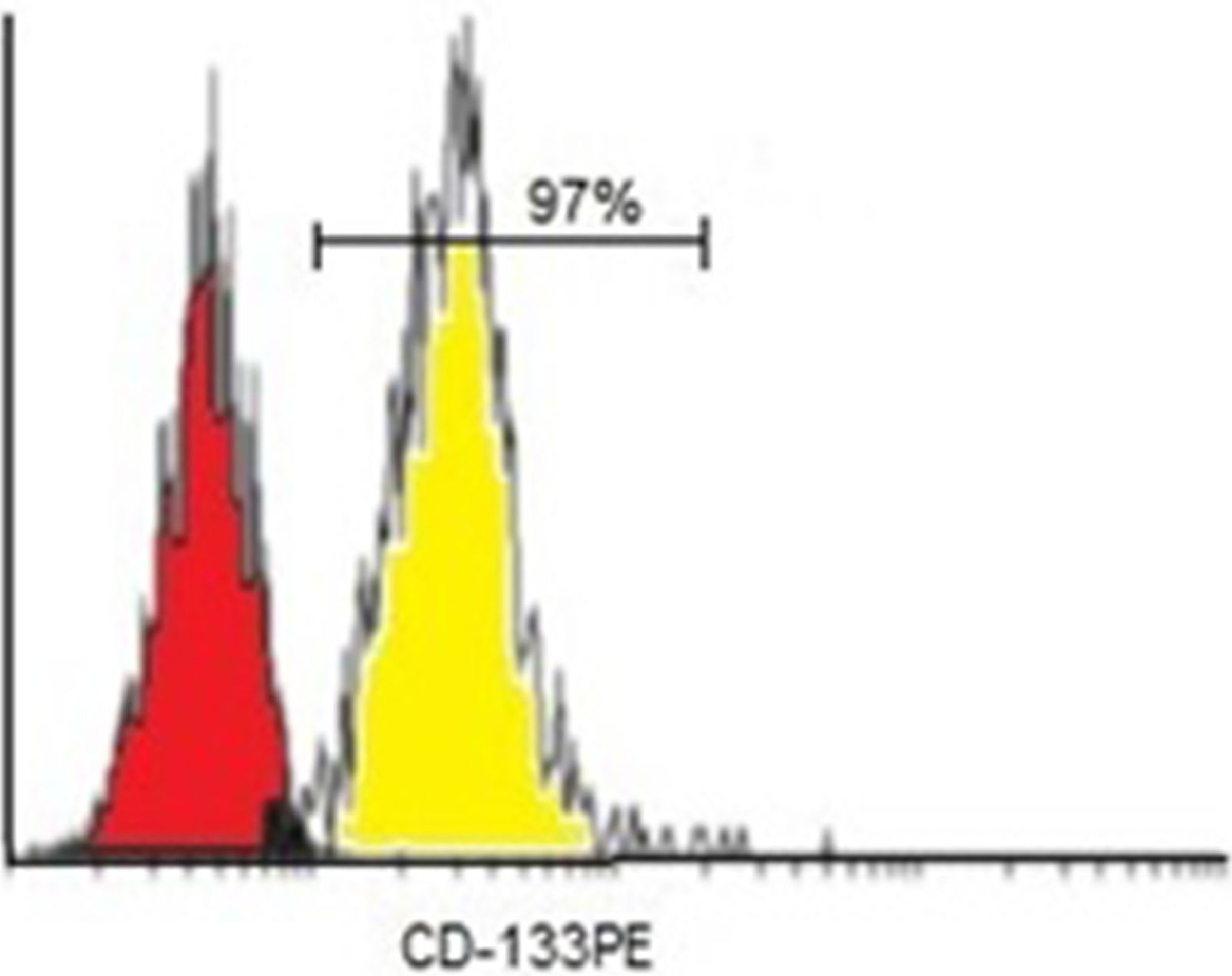
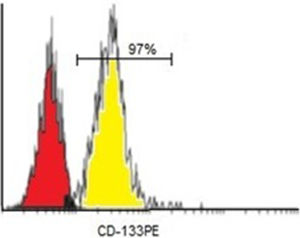
On average, the number of CD133+ cells separated by the MACS was 106. Purity assessments of enriched CD133+ cell fractions were analyzed by a flow cytometer. Flow cytometer analysis after isolation showed that the mean purity of separated cells among samples was 97% for CD133+ cells (Figure 3).
Expansion of CD133+ cells in the tissue culture plates and microfluidic bioreactorThe first goal of this study was to determine the expansion rate of CD133+ cells in microfluidic bioreactors. The CD133+ cells extracted from cord blood were cultured in duplicate in microfluidic devices. At the same time, CD133+ cells were also cultured in 24-well plates for comparison purposes. The expansion of the total nucleated cells (TNCs) and CD133+ cells (n = 5) was calculated. On day 12, the microfluidic system expansion results showed significant improvement, compared to those of the tissue culture plate (TCP), with p < 0.05. The TNCs cultured in the TCP generated a 45-fold expansion, while it expanded 65-fold in the microfluidic system. On day 12, the CD133+ cell expansion was 4.5-fold in the TCP and 8.5-fold in the microfluidic system (Figure 4).
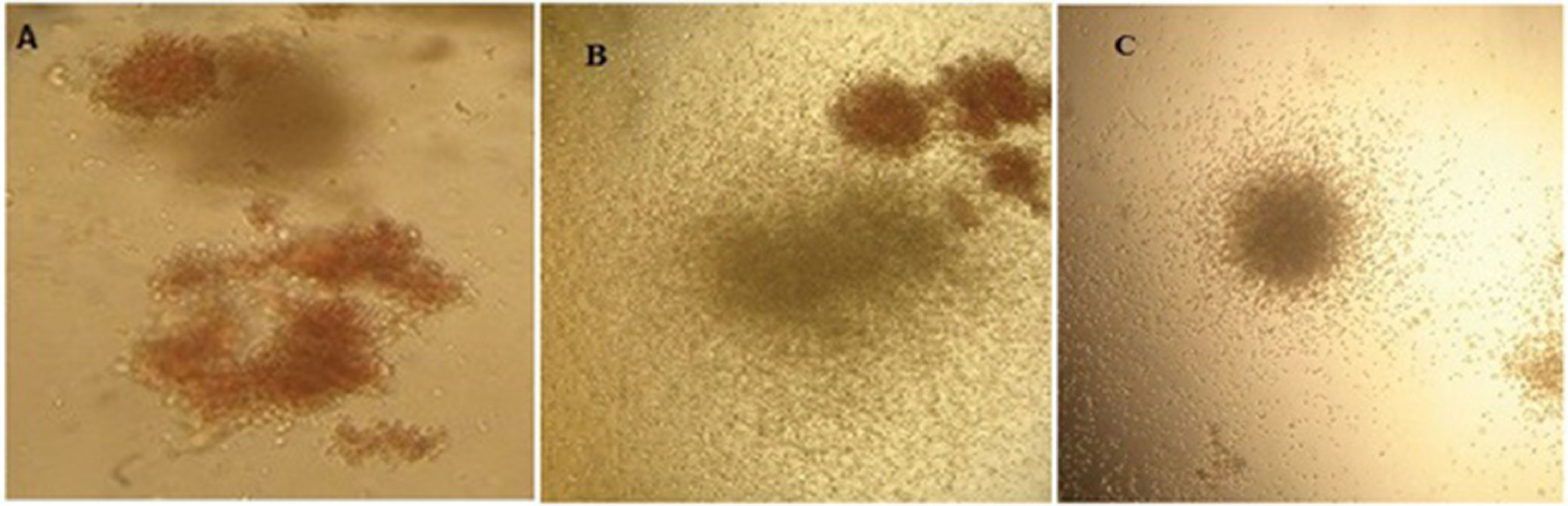
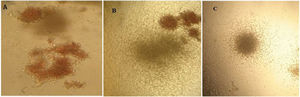
The proliferation of total cells and CD133+ cells. a) TNC cultured in TCP generated 45-fold expansion, while it expanded 65-fold in the microfluidic system (p < 0.05). b) CD133+ cells yielded hematopoietic colonies after 12 days of culture in methylcellulose containing CFU-E, CFU-GEMM and CFU-GM in a microfluidic system (* p < 0.05)
On day 12, expanded cells in the TCP and microfluidic bioreactor were cultured in methylcellulose to compare their colony-forming potential. After 14 days of culturing in methylcellulose, the number of burst-forming unit-erythroid (BFU-E), colony-forming unit-erythroid (CFU-E), granulocyte-macrophage progenitor (CFU-GM) and colony-forming unit – granulocyte, erythrocyte, monocyte and megakaryocyte (CFU-GEMM) colonies were counted by their morphology. It was shown that microcavities of the bioreactor were more efficient in expanding CD133+ cells, but these results were statistically insignificant (p >0.05). This test was repeated three times and, on average, the tissue culture produced 123 colonies and the microfluidic bioreactor produced 149 colonies. The appearance of the colonies is shown in Figure 4.
Assessment of CXCR4 profile of the expanded cellsA critical point of in vitro HSC expansion is the homing ability of these cells and thereby their potential in reviving hematopoiesis. The CXCR4 is a vital factor in such a process. To better understand the role the microfluidic system plays in CXCR4 expression of the expanded hematopoietic cell, we analyzed cell surface expression levels of CXCR4 molecules on CD133+ cells and CXCR4 messenger RNA (mRNA) expression in CD133+ cells cultured in different systems. We first examined the cell-membrane expression of the CXCR4 in the expanded cells by flow cytometry using a phycoerythrin (PE)-conjugated anti-CXCR4. On average, the percentage of freshly purified CD133+ cells (day 0) that were positive for CXCR4 was 77.6% ± 15.2%. Therefore, it was considered to be used as a baseline to study changes in CXCR4 expression during the culture of CD133+ cells. The median percentage of CXCR4 expression in the expanded cells after 12 days was lower in the conventional system, compared to the microfluidic system, with a median expression of 8.9% in the microfluidic system, compared to 7.7% in the conventional system (p < 0.05).
In the second step, in order to assess CXCR4 expression in hematopoietic stem cells, real-time PCR was conducted on RNA extracted from fresh HSC of cord blood and HSC expanded under the conventional and microfluidic system. Threshold cycle values were normalized to the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) expression and the level of CXCR4 expression was calculated relative to the freshly enriched CD133+, as the untreated control. According to our experiment, the CXCR4 gene expression in the microfluidic system remained the same, while expression in the traditional system was reduced. However, the results were not statistically significant (data are not shown). This illustrated the fact that besides surface expression, mRNA levels of CXCR4 were also increased. Thus, the CXCR4 gene is up-regulated in the microfluidic system, compared to the traditional type.
DiscussionOver the past few years, UCB-HSC has created plenty of interest because of its potential uses in clinical HSPC transplantation. In recent years, different strategies have been employed to expand the stem cell population ex vivo, especially those derived from cord blood. In accordance with this, efforts have been made to expand HSCs ex vivo using cytokines and factors governing cell expansion. Unfortunately, we are faced with the challenge of unwanted cell differentiation after expansion.15–17 The CD34 and CD133 are two conventional surface markers for HSCs. In several traditional methods, CD34+ cells are used for expansion purposes, whereas we focused on expanding CD133+ cells that are known to be expressed on less differentiated cells. Danel et al. reported that expanded CD34+ cells had a less severe combined immunodeficiency (SCID)-repopulating capacity and the study by Bonanno et al. concluded that CD133+ cell transplantation has better results than that of CD34+ cells.18,19 In this study, UCB-HSCs were cultured in both tissue culture plates and the bioreactor. Although most of our current knowledge of modern biology is indebted to cell cultures in conventional 2D conditions, in vivo cells are expanded and differentiated in an intricate 3D micro-environment.6,20 For this reason, in an attempt to replicate this milieu, we opted to use the microfluidic structure with microcavities covered with fibronectin that resembles bone marrow niches. Our results showed a 65- and 6-7-fold expansion of TNCs and CD133+ cells, respectively (compared to 45 and 3.5-fold with the traditional method). Flow cytometry displayed a decrease in CD133+ purity from 95% to 9% on day 12. This showcased more expansion and less differentiation for the microfluidic system. Recently, scientists have learned a remarkable change in the structure and function of fibroblasts cultured in 3D conditions. Cukrierman et al. reported a faster expansion of fibroblasts in the 3D environment, compared to that of the 2D.21 Schawrtz et al. demonstrated that gentle perfusion and local oxygenation of hematopoietic cells resulted in greater survival and expansion of progenitors.22 In 2006, Feng et al. cultured HSCs in the polyethylene terephthalate (PET) scaffold coated with FN. Their culture expanded the cells 100-fold in 10 days. Furthermore, their LTC-IC assay experienced a 42-fold increase.23 In our study, we did not witness a significant LTC-IC increase in the traditional or microfluidic device. We also evaluated cell colony-forming potential, but 1.4-fold increase in BFU-E colonies and 1.2-fold increase in total was not statistically significant. Nevertheless, in Feng's experiment, there was a 3.8-fold elevation in colony count. This discrepancy is likely due to different cell sources.
Cell-matrix interaction is another aspect of optimum cell growth. To this end, we covered the microcavity surfaces with FN. The cells bonded with the FN layer and, after 10 days, they proliferated more than those in the traditional culture. A study by Jiang et al. concluded that FN is not only an anchor for stem cells, but it also affects proliferation.24 In an experiment by Hurley et al., HSCs neighboring FN expressed the CD133+ marker at a higher level than those without FN.25
A robust chemoattractant for HSCs produced by bone marrow stromal cells is the SDF-1 (CXCL12) and its receptor on the HSC, CXCR4, which are crucial in the BM homing of hematopoietic stem cells to the point that loss of the CXCR4 could lead to impaired homing. Yong-Rui Zou demonstrated that impaired hematopoiesis in CXCR4−/− mice is indistinguishable from that of SDF-1−/- mice.26 In our study, we assessed the expression of the CXCR4 and its mRNA. Expression of the CXCR4 marker had a prominent decrease after 12 days of expansion, with 7.7% and 8.9% in traditional and microfluidic conditions, respectively. Our results are vindicated by several research groups claiming that the UCB-HSC in vitro culture, which is needed for HSPC transplantation, diminishes the CXCR4 expression and only a small fraction of culture-expanded HSCs are CXCR4 positive.27,28 Finally, mRNA levels of CXCR4 in the 2D culture experienced a drop, but this was not the case in microfluidic-expanded cells. The CXCR4 surface expression was not in agreement with its mRNA consistent expression. This could be due to faulty protein production in ex vivo-expanded cells or the engulfed surface CXCR4. Our study showed that massive ex vivo expansion of UCB-HSCs in fibronectin-coated micro-cavities of the microfluidic bioreactor is feasible. This construct was used for the purpose of expanding HSCs with a higher colony-forming capacity, which can maintain more CXCR4 molecules, in order to increase overall expansion, compared to conventional 2D cultures.
ConclusionOur findings provide a basis for the microenvironment matrix constitutive. However, we suggest additional studies be performed to observe the effect of coating with a combination of matrix proteins, such as collagen, laminin and fibrinogen.
We are grateful to Yan Zhou, Xu Zhang, Minglong Zhu and Ping Hua for their help in the laboratory and Lin Wei and Dongning Pan for their work on the cDNA. We would also like to thank Zhihua Huang and Ping Hu for their instructive discussion and help.