The Kidd system (e.g. JK; ISBT0091) is the ninth blood group system, which was identified between 1951 and 1959 following a case of hemolytic disease of a newborn (JK1 in 1951) and hemolytic reactions after blood transfusions (JK2 in 1953 and JK:−1,−2 in 1959) by Allen et al.,2 Plaut et al.3 and Pinkerton et al.,4 respectively. The JK gene (SLC14A1 or HUT11 or UT-B1) is located on chromosome 18q11–q12 with 10 exons, exons 4–10 of which are responsible for encoding the mature protein. It functions as a urea transporter on erythrocytes as well as on endothelial cells of the vasa recta, protecting the erythrocytes from osmotic lysis and allowing urine concentration in the kidneys. The Kidd system includes three antigens (JK1, JK2 and JK3); JK1 and JK2 are antithetic, JK3 is always present when JK1 and/or JK2 are/is present. This system leads to four RBC phenotypes (JK:1,−2; JK:1,2; JK:−1,2 and, more rarely, JK:−1,−2).1–5
The Kidd system is one of the most immunogenic systems in human red blood cells (RBCs), associated with acute and delayed hemolytic reactions. Although immunization is rare (< 1 %), alloimmunization against all JK antigens may occur due to JK system incompatibility following an immunization event (e.g. transfusion) or to an unknown stimulus (transfusions in figures at the CHU UCL Namur in 2020: 11,468 RBC units transfused; 18,107 IAS [irregular antibody screening] realized; combined anti-JK1 and JK2 incidence = 0.05 %; these data have not yet been published). Although alloantibodies against absent JK antigens have already been well investigated, the literature on alloantibodies against JK antigen variants is scarce, which results in limited knowledge regarding conflicts between the JK1 and JK2 phenotypes and the concurrent respective alloantibodies.(1, 2) It is worth noting that naturally occurring anti-JK1 without obvious stimuli have been reported in an infant and in a healthy blood donor.8–10 In this study, we report a complex case of a contradictory anti-JK1 in the presence of JK1 phenotype in a patient who had previously received JK1 RBCs. We also highlight the pitfalls of multiple routine methods in such cases, and show that new approaches such as molecular genotyping of RBCs are more useful and reliable in understanding and explaining these discrepancies.
Case reportIn November 2018, a 57-year-old male patient was admitted to our institution with chronic anemia due to multiple deep pressure sores. Confirming the anemia by laboratory results (Table 1), internists requested two red blood cell units (RBCUs) to treat the patient. The initial double red blood cells (RBCs) phenotype determination was O RH:1,2,−3,4,5; KEL:−1,2. Following a negative irregular antibody screening (IAS; Ortho BioVue system, NJ, USA), the patient received two RBCUs consisting of (1) O RH:1,2,−3,4,5; KEL:−1,2; FY:1,2; JK:1,2; MNS:3,4; and (2) O RH:1,2,3,4,5; KEL:−1,2 (unknown extended phenotype). For the same reason, the patient was readmitted to our hospital in January 2022. Unlike the first time, the automatized IAS emerged as positive. Therefore, we performed more extensive RBC phenotyping and direct and indirect antiglobulin tests (DAT and IAT). Further determination of the patient's RBC phenotype using a serological assay (Ortho BioVue system) was as follows: RH:2,−3,4,5 (reaction intensity: 4+/0/4+/4+), KEL:−1,2 (0/4+), JK:1,2 (3+/3+), MNS:1,2 (3+/2+). There was positive interference (positive auto-control cells) with FY1, FY2, MNS3 and MNS4 antigens that use the immunoglobulin (Ig)G reagents for antigen typing of RBCs. Thus, we did not interpret the results for the latter antigens. The DAT was interpreted as slightly positive (1+). Identification of anti-erythrocyte antibodies was performed on the Orthovision analyzer (Ortho Clinical Diagnostics, NJ, USA) using two sets of 11 microcolumns (Ortho BioVue System) prefilled with commercial erythrocytes of known phenotype (0.8 % Resolve panel C-untreated and Ficin-treated; Ortho). The antigram antigen profile (Ortho) allowed technicians and clinical pathologists to accurately identify allo-anti-RH3 (because of a previous transfusion with RH3 RBCs). After a negative pretransfusion compatibility test, the patient received one O RBCU with RH:1,2,−3,4,5; KEL:−1,2; FY:1,2; JK:1,−2 and MNS:3,−4 phenotype. RBC genotyping was not performed at the second admission.
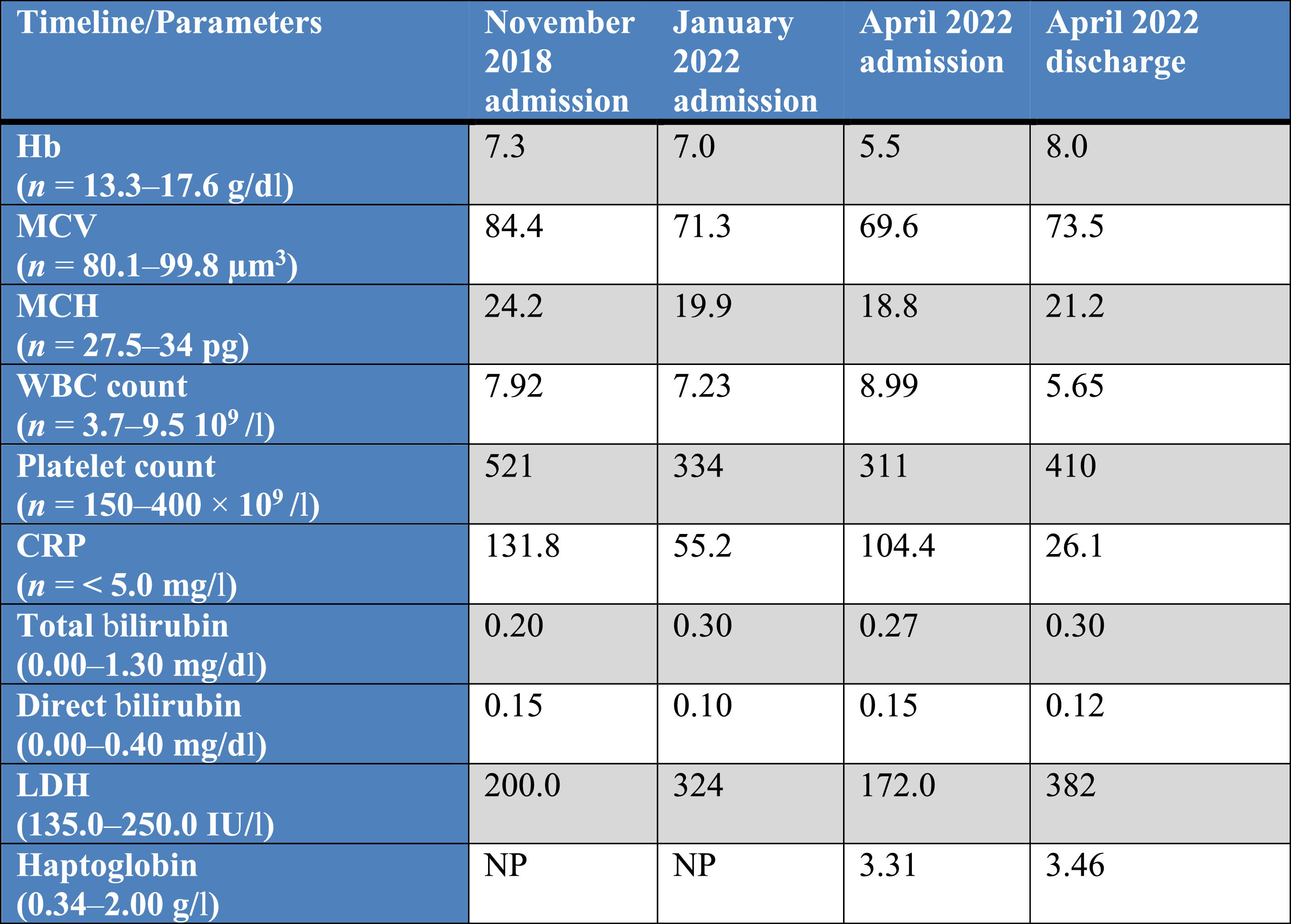
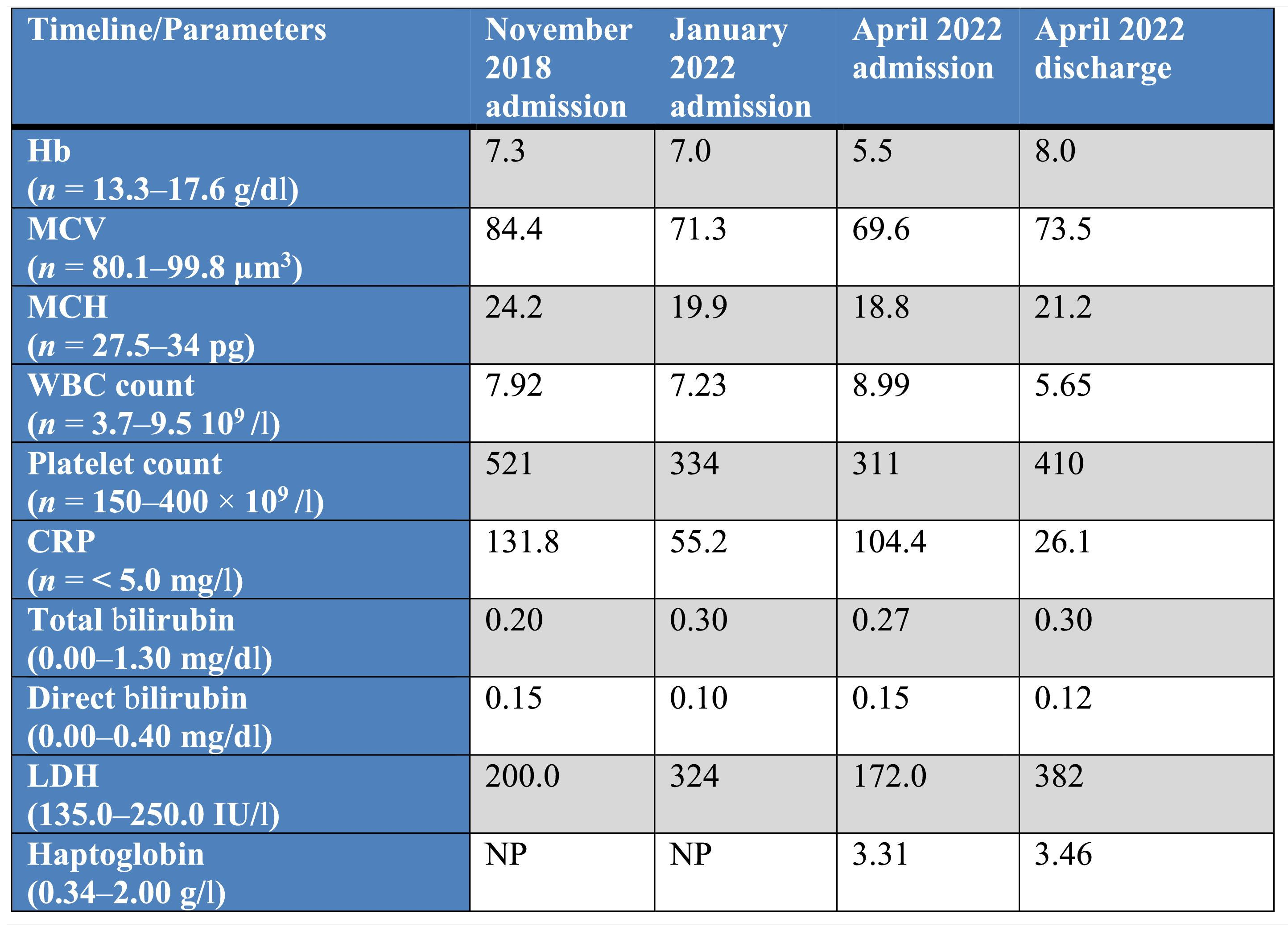
Laboratory assessment on admission and before each blood transfusion and after the last discharge, between 2018 and 2022.
Abbreviations: Hb: hemoglobin; MCV: mean cell volume; MCH: mean cell hemoglobin; WBC: white blood cell count; CRP: C-reactive protein; LDH: lactate dehydrogenase; NP: not performed.
On the last admission in April 2022, we received an order of two RBCUs for the same reason at the transfusion service and donor blood center. On admission, the patient was normothermic, normotensive and had normal heart and respiratory rates. Laboratory results at each admission from 2018 to 2022 are summarized in Table 1. He was polymedicated, but had not taken any documented medications correlated with auto-anti-RBC production.11
As IAS was positive and showed interference in the auto-control cell, we performed DAT and IAT tests following our procedures. The DAT emerged as positive for both IgG (reaction intensity: 3+) and C3b–C3d (2+) reagents. Identification of anti-erythrocyte antibodies, including anti-JK, was performed as previously explained. The antigram antigen profile allowed us to identify anti-RH3 for the second time, as well as the new anti-JK1. As the latter result was not consistent with the patient's RBC JK:1,2 phenotype and the auto-control cell was also positive (interfering factor), we retested the patient's plasma with a different batch of Ortho reagent (lots 8RC380 and 8RC382). To confirm our observations, we also used another commercial set of 11 untreated microcolumns prefilled with commercial erythrocytes of known phenotype for antibody identification (ID-DiaPanel; BioRad, CA, USA). An acid elution process was carried out (DiaCidel; BioRad) to identify RBC-bound antibodies, which also revealed the presence of anti-RH3 and anti-JK1. The treatment of the patient's plasma with 1,4-dithiothreitol (DTT; Merck KGaA, Darmstadt, Germany) revealed the presence of warm IgG antibodies to RH3 and JK1 antigens. We repeated all the tests on a second collected plasma sample 4 days later. The second results confirmed the initial observations. Given the interference with IgG-positive DATs (e.g. medicine, auto-antibody, etc.) and conflicting phenotyping, we used the single specific primer–polymerase chain reaction (SSP–PCR) technique on a commercial kit; namely, inno-train RBC-Ready Gene (inno-train Diagnostik GmbH, Hesse, Germany), to confirm the patient's RBC phenotyping. Extraction of the DNA was carried out manually with the QIAamp DNA Blood Mini Kit (Qiagen, Venlo, the Netherlands) and amplification was performed with an ABI 9700 thermal cycler (ThermoFisher Scientific, MA, USA) according to the manufacturer's instructions. The amplification parameters were as follows: one cycle at 94 °C for 2 min, followed by five cycles consisting of two steps: (1) at 90 °C for 20 s and (2) at 70 °C for 60 s, followed by 10 cycles of three steps: (1) at 94° for 20 s, (2) at 65 °C for 60 s and (3) at 72 °C for 45 s, followed by 20 cycles of three steps: (1) at 94 °C for 20 s, (2) at 61 °C for 60 s and (3) at 72 °C for 45 s; and finally, an extension step at 72 °C for 5 min. The amplification products were then electrophoresed at 140 Vs for 8 min on a 3 % agarose gel using a tris–borate–ethylenediamine tetraacetic acid (EDTA) buffer solution containing a 10,000 × fluorescent DNA gel stain (SYBR Safe DNA gel stain; Invitrogen, Paisley, UK) and visualized using blue-light illumination. The results were then compared with different standard molecular weights to identify the RBC genotypic pattern, which confirmed the predicted phenotype while providing more information regarding other antigens. The genotypic profile was as follows: RH:1,2,−3,4,5; KEL:−1,2; FY:−1,2; JK:1,2; MNS:1,2,3,−4; DO:1,2; VEL:1.
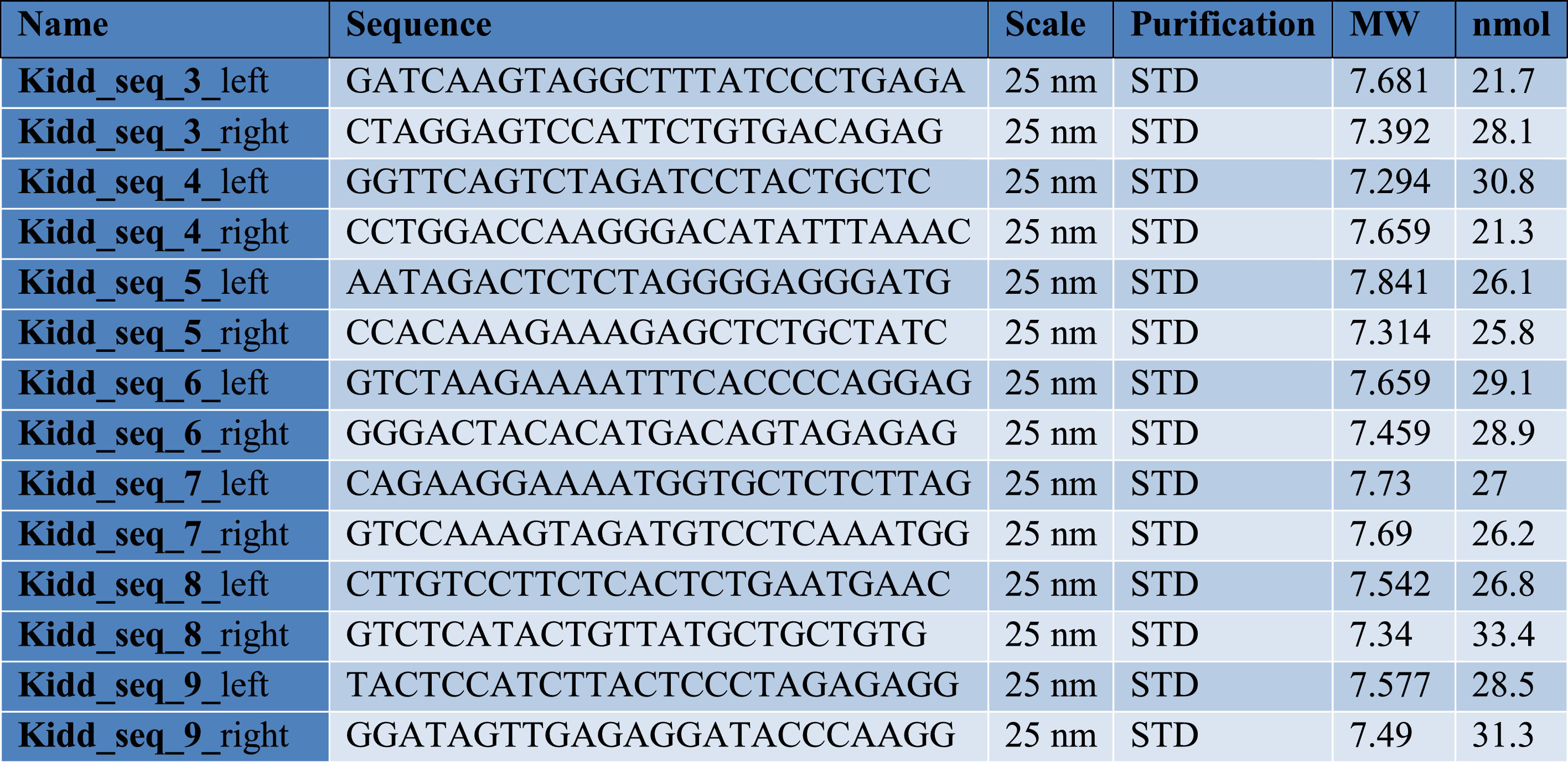
Finally, to more clearly understand the discrepancy between RBC phenotype and antibody presence, we genotyped the patient's RBCs using a newly designed in-house gDNA sequencing method for RBC typing with long-read sequencing technology. For this purpose, we used amplicon-based sequencing relying on a set of seven newly developed primer pairs that spanned the whole SLC14A1 gene. Whole blood DNA extraction was performed with the MagAttract HMW DNA kit (Qiagen). Primer pairs (see Table 2) were used separately and a PCR was performed per pair with a QuantStudio™ 5 real-time PCR System (ThermoFisher). PCR products were then pooled and prepared for sequencing, using the rapid barcoding kit from Oxford Nanopore Technologies (Oxford, UK; SQK-RBK 110–96), according to the manufacturer's instructions. Barcoded samples were pooled to form a sequencing library and purified by solid-phase reversible immobilization using magnetic beads. The purified library was loaded onto an R9.4 flow cell (Flo-Min 106; Oxford Nanopore Technologies) and sequencing was performed on a GridION (Oxford Nanopore Technologies) platform in super-accurate base-calling mode. Real-time base-calling and demultiplexing of raw data were carried out using MinKnow (version 22.05.7), generating FASTQ files. These files were processed through an in-house bioinformatic pipeline, including assembly, alignment and variant calling tools.
Whole genome sequencing of the JK locus showed a c.838G>A (rs2298720) transition, confirming the heterozygosity of the JK gene (variant allele frequency of 52 % for JK*01 and 48 % for JK*02). We also identified two significant unique single nucleotide variants (SNVs), including the heterozygous c.130G>A (rs1058396) and the homozygous c.588A>G (rs2298718) (see Figure 1), based on the ISBT009 JK blood group alleles version 8.0 data sheet.12 The combination of c.130G>A and c.588A>G SNVs on the JK*01 allele, corresponding to the JK*01W.06 allele, appears to be responsible for the low expression of the JK1 antigen (see Figure 1). Therefore, the patient's whole SLC14A1 genotype was found to be JK*01W.06/JK*02. Based on the serological phenotyping, anti-RBCs screening, genotyping result and clinical data, we presume that the detected anti-JK1 could be an alloantibody developed after transfusions of RBCU with the JK1 phenotype.
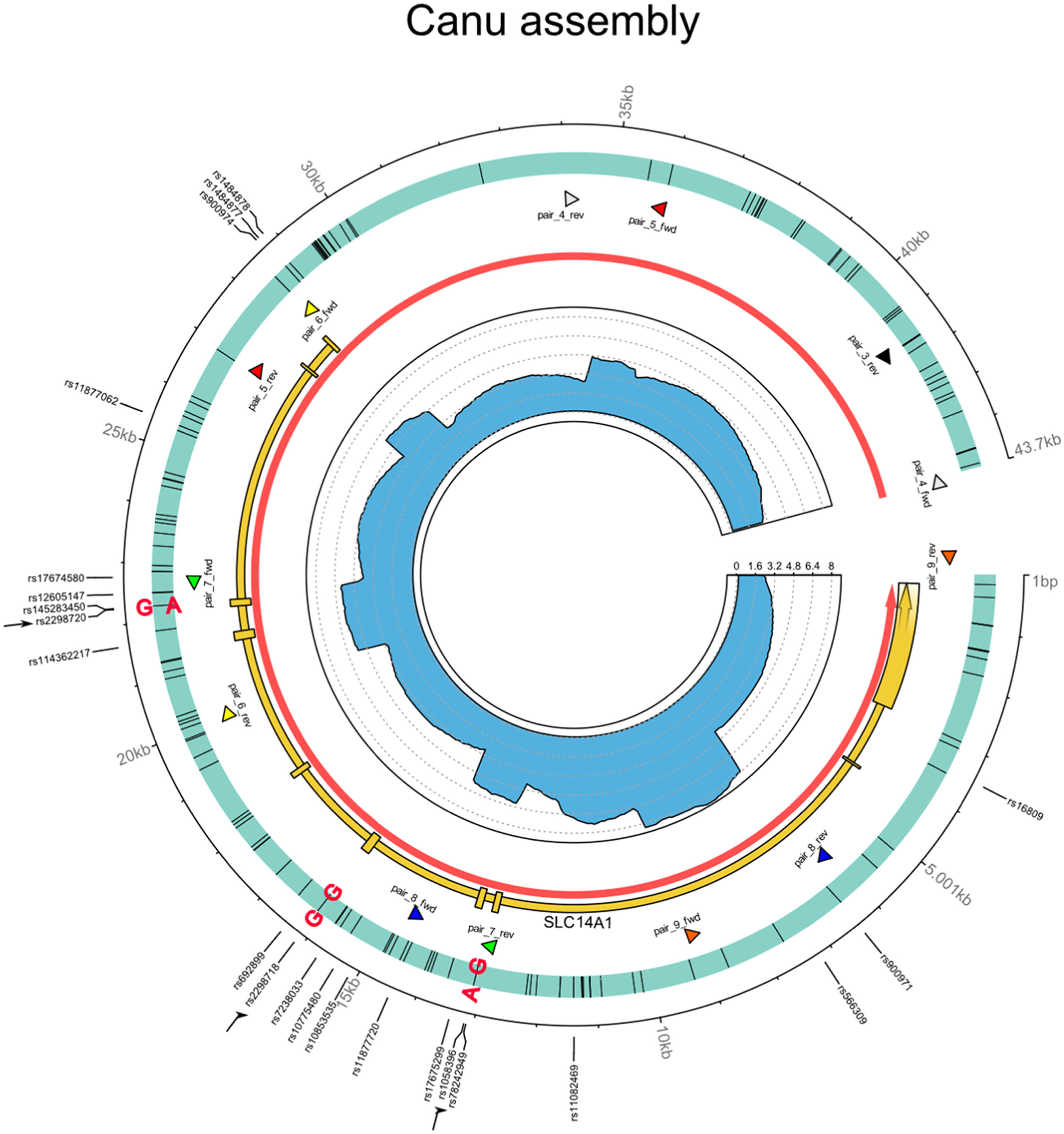
Circos plot of the SLC14A1 gene using Canu for genome assembly, illustrating the sequencing results and allelic distribution of the mutations.
Circos plot of the de-novo assembled region covering SLC14A1 gene (Homo sapiens, chr 18: 45 719 929 to 45 756 770). The outer track indicates contig coordinates (in bp) and the sites with known polymorphism according to the Reference SNV report (rs) NCBI. For convenience, the three sites of major importance here are indicated by black arrows. The second track represents the assembled contig with all SNVs (black bars) compared to the reference NC00018 and relevant mutations are indicated in bold type; each phased allele is presented separated by “|”. The third track shows the location of the SLC14A1 (MANE project release version 1.0; accession NM_015865.7) gene in yellow with exons represented by boxes, the location of the SLC14A1 gene (according to Ensembl release 109 in red; accession ENSG0000141469), as well as the primer pairs (arrow heads, each pair with its own color) defined in the present study. The inner track highlights the sequencing coverage (divided by 1000 for clarity).
As we did not have an RBCU that completely matched the patient's genotype, and after negative pretransfusion compatibility tests, we delivered two compatible JK:−1,2 RBCUs with confirmed phenotypes: (1) O, RH:1,2,−3,4,5; KEL:−1,2; FY:1,2; JK:−1,2; MNS:3,4; and (2) O, RH:1,2,−3,−4,5; KEL:−1,2; FY:1,2; JK:−1,2; MNS:−3,4. The patient was discharged a week later with no evidence of hemolytic reaction (Table 1) and to date, according to his medical record, has not received another RBCU.
DiscussionIn this study, we have presented the clinical case of a patient with JK:1,2 RBC phenotypes, concomitant allo-anti-JK1 and the JK*01W.06 polymorphism. Although this allele is not new, it is probably the origin of the development of allo-anti-JK1 after JK1 RBC transfusions, which makes the case interesting. Indeed, the JK1 antigen is the third immunogenic antigen after RH1 and KEL1 compared to other blood group antigens to induce alloantibodies in transfused populations. They are known to be perfidious and dangerous, as their titers often fall below the detection threshold weeks or months after an immunization event. Besides acute reaction, anti-JK are a very common cause of delayed hemolytic reactions.1,6,13 In this case, we argue that the first alloimmunization occurred during the fall of 2018 after JK1 RBC transfusions. Anti-JK1 titers dropped over time and became undetectable on our serological assay in January 2022 (second admission). Therefore, we presume that the anamnestic reactivation in April 2022 was due to another transfusion of a RBCU with JK1 phenotype. As all the auto-control cells were positive and the DAT, DTT procedure and acid elution treatment revealed the presence of IgG antibodies directed against the JK1 antigen, we could not formally exclude the presence of auto-anti-JK1. However, knowing that transfused RBCs usually have a reduced lifespan (approximately 90 days), and based on phenotypic and genotypic results, as well as clinical data including the chronology of immunization events, we presume that the patient had developed earlier allo-anti-JK1 against the more common structural form of JK1 antigen on transfused RBCs.
Sequencing of the whole SLC14A1 gene revealed two significant unique SNVs (see above), suggesting the presence of the JK*01W.06 weak allele. The c.130G>A SNV is responsible for weak JK1 antigen expression when identified alone and is associated with the JK*01W.01 allele, whereas c.588A>G is commonly associated with the JK*02 allele. However, when c.130G>A combines with c.588A>G, the associated allele is JK*01W.06.12 We therefore conclude that the patient's SLC14A1 genotype is JK*01W.06/JK*02 and his phenotype is Jk1w,2. However, our serological test (Ortho) was not affected by the low expression of JK:1 antigen and showed a reaction intensity of 3+/3+ for both JK:1,2. These results are normal and do not suggest the low expression of JK antigens in our daily routine. Further reports are needed to confirm such discrepancies.
The more prominent SNV, which is a c.130G>A missense variant (leading to weak phenotype JK*01 W.01), has been well described in the literature with normal or weakened agglutination pattern.6 Allo- and autoantibodies associated with JK*01W.01 allele have also been reported previously.6,7 However, when combined with the c.588A>G silent variant resulting in the JK*01W.06 allele, the literature appears scarce and our knowledge is limited. As the c.588A>G is a silent variant, we would not claim that it has an impact on the protein structure. Although we acknowledge that the literature on this topic is scarce, it could be argued that the c.588A>G variant introduces a new CpG motif into the DNA sequence which might lead to methylation of the cytosine residue and therefore impact the expression of the SLC14A1 gene. Wester et al. has stated previously that c.588A>G silent variant, like many other silent variants, may contribute to a weak expression of JK antigens by affecting the amount of mature protein.14
We also observe the clinical significance of these alloantibodies, as the patient's hemoglobin decreased drastically in April 2022, 3 months after the JK1 RBC transfusions in January 2022. As previously stated, although we cannot formally rule out the presence of auto-anti-JK1, our findings are consistent with alloantibodies directed against the JK1 antigen. Indeed, various routine methods (IAS, IAT, DAT, acid elution, etc.) used in this case cannot answer the latter question clearly, showing the complexity of the concomitant JK1 phenotype associated with anti-JK1 presence. Furthermore, all our routine methods are affected by the presence of interferences (e.g. complement, IgG antibodies). One limitation of our study is the lack of a more effective serological assay to clarify and confirm whether anti-JK1 was an auto- or an alloantibody. As auto-anti-JK1 have been described in the context of autoimmune diseases, chemical substance intoxications or viral infections,15–21 and as we cannot find any known probable stimuli related to the detected antibodies except for previous blood transfusions with JK1 RBCs, we rule out the presence of auto-anti-JK1. Thus, we classify the anti-JK1 detected in the patient's plasma as alloantibodies.
In the meantime, we suggest multiple approaches and more reliable methods such as PCR or sequencing to describe immunized cases with conflicting profiles. We recognize that these methods are expensive, time-consuming, laborious and require skill, but as there is no specific commercial serological assay we recommend that patients presenting such a profile are transfused with JK:−1 RBCs to avoid any hemolytic reaction, even with a negative antibody screening or pretransfusion compatibility.
In this study, we show the complexity of treating cases such as the one elaborated here which requires close collaboration between the patient, clinicians, hospital blood transfusion service and the clinical laboratory.