In acute lymphoblastic leukemia (ALL), remission is classically defined as the reestablishment of normal hematopoiesis and the presence of less than 5% of the nucleated blast cell population found by conventional microscopy; this is used in older protocols to assess treatment response. Morphological analysis, although useful and applicable at any center, has proven to be of limited sensitivity, subjective and imprecise to study early response to treatment and this technique does not appear to be sufficient to identify patients at true risk of relapse who might benefit from the intensification of treatment.1,2 For this reason, cytomorphological analysis has been replaced by minimal residual disease (MRD) monitoring in several treatment protocols and new definitions of remission and relapse in childhood ALL have been proposed.3
The analysis of MRD has proved to be the strongest independent prognostic factor in all studies analyzing large series of patients with B-lineage and T-cell ALL, and specific molecular subgroups such as patients with the BCR-ABL fusion gene and ALL patients with MLL gene rearrangements. This analysis allows more accurate risk group assignment and tailoring the intensity of treatment, permitting reduction or intensification at the different treatment time points according to the MRD level.4–9 MRD monitoring can also guide treatment decisions in relapsed patients and those who are candidates for bone marrow transplantation.4,5,10,11
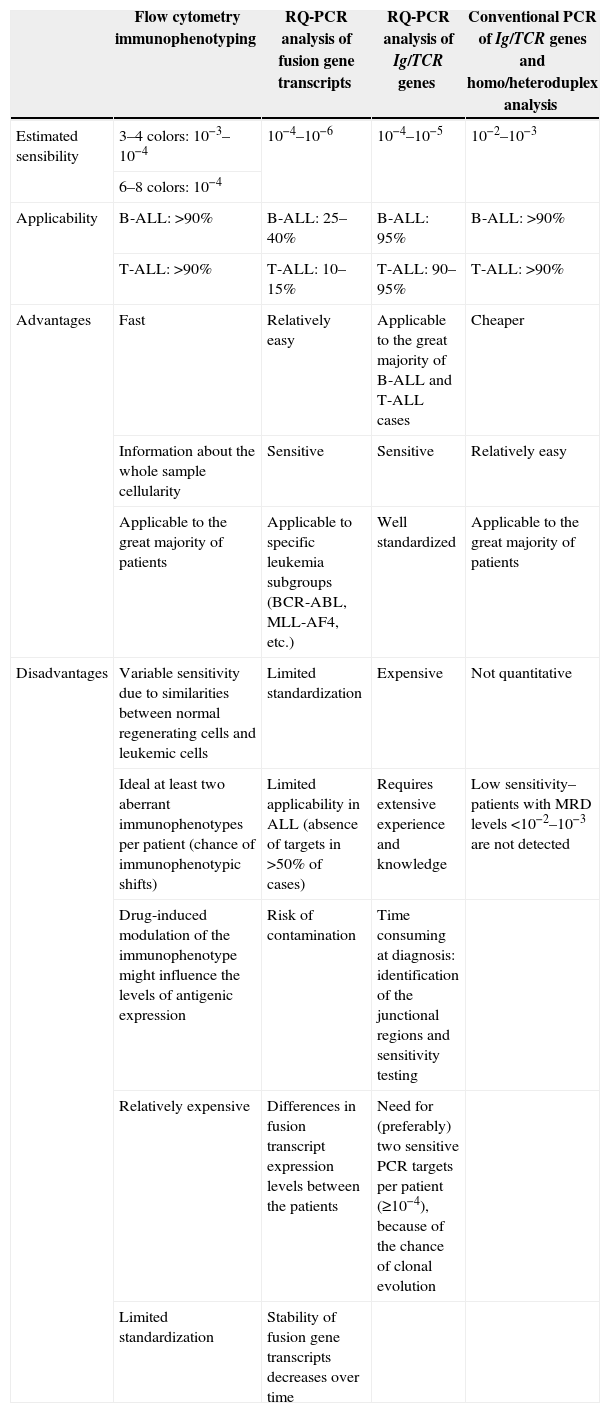
Sequential monitoring of MRD using more sensitive and specific techniques, such as quantitative real-time polymerase chain reaction (RQ-PCR) for immunoglobulin (Ig) and T-cell receptor (TCR) gene rearrangements and flow cytometry analysis, with a detection power of one blast cell in 104–106 normal cells, has substantially refined the assessment of early response to treatment. Unfortunately these methods are not only expensive, but technically complex and require considerable technology and highly-specialized laboratories to be routinely used in risk stratification protocols for ALL; they are therefore inaccessible to most treatment centers, especially in developing countries.4,12 The development of simplified MRD technologies is essential to allow the potential benefits of MRD monitoring to be extended to all children with leukemia including those treated in low-budget countries. Table 1 shows some characteristics of the main methodologies used to detect MRD in ALL.
Characteristics of the methodologies used for minimal residual disease detection in acute lymphoblastic leukemia (ALL)a.
| Flow cytometry immunophenotyping | RQ-PCR analysis of fusion gene transcripts | RQ-PCR analysis of Ig/TCR genes | Conventional PCR of Ig/TCR genes and homo/heteroduplex analysis | |
|---|---|---|---|---|
| Estimated sensibility | 3–4 colors: 10−3–10−4 | 10−4–10−6 | 10−4–10−5 | 10−2–10−3 |
| 6–8 colors: 10−4 | ||||
| Applicability | B-ALL: >90% | B-ALL: 25–40% | B-ALL: 95% | B-ALL: >90% |
| T-ALL: >90% | T-ALL: 10–15% | T-ALL: 90–95% | T-ALL: >90% | |
| Advantages | Fast | Relatively easy | Applicable to the great majority of B-ALL and T-ALL cases | Cheaper |
| Information about the whole sample cellularity | Sensitive | Sensitive | Relatively easy | |
| Applicable to the great majority of patients | Applicable to specific leukemia subgroups (BCR-ABL, MLL-AF4, etc.) | Well standardized | Applicable to the great majority of patients | |
| Disadvantages | Variable sensitivity due to similarities between normal regenerating cells and leukemic cells | Limited standardization | Expensive | Not quantitative |
| Ideal at least two aberrant immunophenotypes per patient (chance of immunophenotypic shifts) | Limited applicability in ALL (absence of targets in >50% of cases) | Requires extensive experience and knowledge | Low sensitivity–patients with MRD levels <10−2–10−3 are not detected | |
| Drug-induced modulation of the immunophenotype might influence the levels of antigenic expression | Risk of contamination | Time consuming at diagnosis: identification of the junctional regions and sensitivity testing | ||
| Relatively expensive | Differences in fusion transcript expression levels between the patients | Need for (preferably) two sensitive PCR targets per patient (≥10−4), because of the chance of clonal evolution | ||
| Limited standardization | Stability of fusion gene transcripts decreases over time |
PCR: polymerase chain reaction; RQ-PCR: quantitative real time polymerase chain reaction; Ig: immunoglobulin gene; TCR: T-cell receptor gene; B-ALL: B-lineage ALL; T-All: T-cell ALL.
A clinically useful simplified MRD technique should be economically viable, widely applicable, specific and sensitive enough to predict the course of the disease. The detection of clonal Ig and TCR rearrangements by PCR and homo-heteroduplex analysis has proved to be a rapid and much simpler and cheaper method than the use of clone-specific probes or flow cytometry. In a multicenter retrospective study, this was the strongest independent prognostic factor in patients treated according to the Grupo Brasileiro de Tratamento da Leucemia Infantil-leucemia linfoide aguda protocol 1999 (GBTLI-99).13 This method represents a good predictive criterion of unfavorable course in children with ALL as it is able in identify patients with a high risk of relapse. This method, however, was not truly quantitative and, due to its lower sensitivity, it should be employed only to identify patients with a high residual tumor load.
Actually in the GBTLI-2009 protocols, MRD analysis at Days 14 and 35 of the induction phase has been used to stratify patients as good and poor responders, guiding treatment decisions in all pediatric ALL subtypes.14 Due to the cost and technical complexity, MRD analysis using patient specific probes by RQ-PCR has been routinely used in very few treatment centers in Brazil and no comparison of this method with simplified MRD strategies to detect Ig or TCR clonal rearrangements by conventional PCR and homo-heteroduplex analysis has been published until now.
In this issue of the Revista Brazileira de Hematology e Hemoterapy, Paula et al.15 compared MRD monitoring using Ig and TCR gene rearrangements by conventional PCR followed by homo-heteroduplex analysis with clone-specific probes to RQ-PCR at the end of induction in 44 children with ALL. According to RQ-PCR MRD cut-off points established by the GBTLI-2009 protocol, the agreement between the two methods was 40% for B lineage ALL and 100% for T-cell ALL. MDR detection by the simplified method was a significant prognostic factor for 3.5-year leukemia free survival. Surprising, the same was not observed using the clone-specific RQ-PCR method. Despite the deficiencies associated with the study design, especially the relatively small number of patients analyzed, the short follow up and the different protocols used – which are well recognized by the authors – the results are interesting and can be useful to aid the validation of alternative and cost effective methods to detect MRD in centers with lower technological resources. Analysis of a larger series of patients with ALL using the same protocol is essential to define the real utility of this simplified strategy in the treatment stratification.
Conflicts of interestThe authors declare no conflict of interest.
See paper by Paula et al. on pages 373–80.