Acute myeloid leukemia presenting the MYST3-CREBBP fusion gene is a rare subgroup associated with hemophagocytosis in early infancy and monocytic differentiation. The aim of this study was to define the relevant molecular cytogenetic characteristics of a unique series of early infancy acute myeloid leukemia cases (≤24months old), based on the presence of hemophagocytosis by blast cells at diagnosis.
MethodsA series of 266 infant cases of acute myeloid leukemia was the reference cohort for the present analysis. Acute myeloid leukemia cases with hemophagocytosis by blast cells were reviewed to investigate the presence of the MYST3-CREBBP fusion gene by fluorescence in situ hybridization (FISH) and reverse transcription polymerase chain reaction.
ResultsEleven cases with hemophagocytosis were identified with hemophagocytic lymphohistiocytosis being ruled out. Six cases were classified as myelomonocytic leukemia, three as AML-M7 and two as AML-M2. In five cases, the presence of the MYST3-CREBBP fusion gene identified by molecular cytogenetics was confirmed by fluorescence in situ hybridization. All patients received treatment according to the Berlin–Frankfürt–Münster acute myeloid leukemia protocols and only one out of the five patients with the MYST3-CREBBP fusion gene is still alive.
ConclusionsOur findings demonstrate that the presence of hemophagocytosis in acute myeloid leukemia was not exclusively associated to the MYST3-CREBBP fusion gene. Improvements in molecular cytogenetics may help to elucidate more complex chromosomal rearrangements in infants with acute myeloid leukemia and hemophagocytosis.
Distinct cytogenetic subgroups of acute myeloid leukemia (AML) have been associated with age-specific frequencies and the incidence of unbalanced aberrations; in particular complex karyotypes increase sharply with age.1 AML presenting the reciprocal translocation (8;16)(p11;p13) that generates the MYST3-CREBBP (former named as MOZ-CBP) fusion gene is mostly observed in adult patients.2 The fusion of the MYST3 and CREBBP genes occurs when both show histone acetyltransferase activities leading to the activation of several targets involved in transcriptional regulation and cell cycle control.2–4 The evidence of AML with the MYST3-CREBBP fusion gene in children was reported by the International Berlin–Frankfurt–Munster (I-BFM) study group.5 Sixty-two pediatric AML were identified in which karyotype records revealed t(8;16)(p11;p13) in the AML observed at an early age, monocyte differentiation [French–American–British classification (FAB) AML-M5] and presence of hemophagocytosis; all of which are associated with very poor outcomes.5 Furthermore, the MYST3-CREBBP fusion gene associated with disseminated intravascular coagulation and high mortality rates was observed in a series of French AML patients.6 These particular clinical, cytological, cytogenetic, and molecular characteristics of AML with MYST3-CREBBP led to the suggestion of a unique category in the World Health Organization (WHO) classification due to the poor prognosis.7 Among the clinical spectrum conditions, hemophagocytic lymphohistiocytosis (HLH) should be included as differential diagnosis. However, HLH presents phagocyte activation caused by immune disorders that compromise T cell/natural killer cells and the normal monocyte-macrophage lineage.8
An accurate case identification requires the evaluation of morphological, cytogenetic and molecular features following correlation of obtained parameters, including serological tests. In this study, the availability of a unique series of early onset AML cases prompted us to search for AML-MYST3-CREBBP cases and to define relevant molecular cytogenetic characteristics.
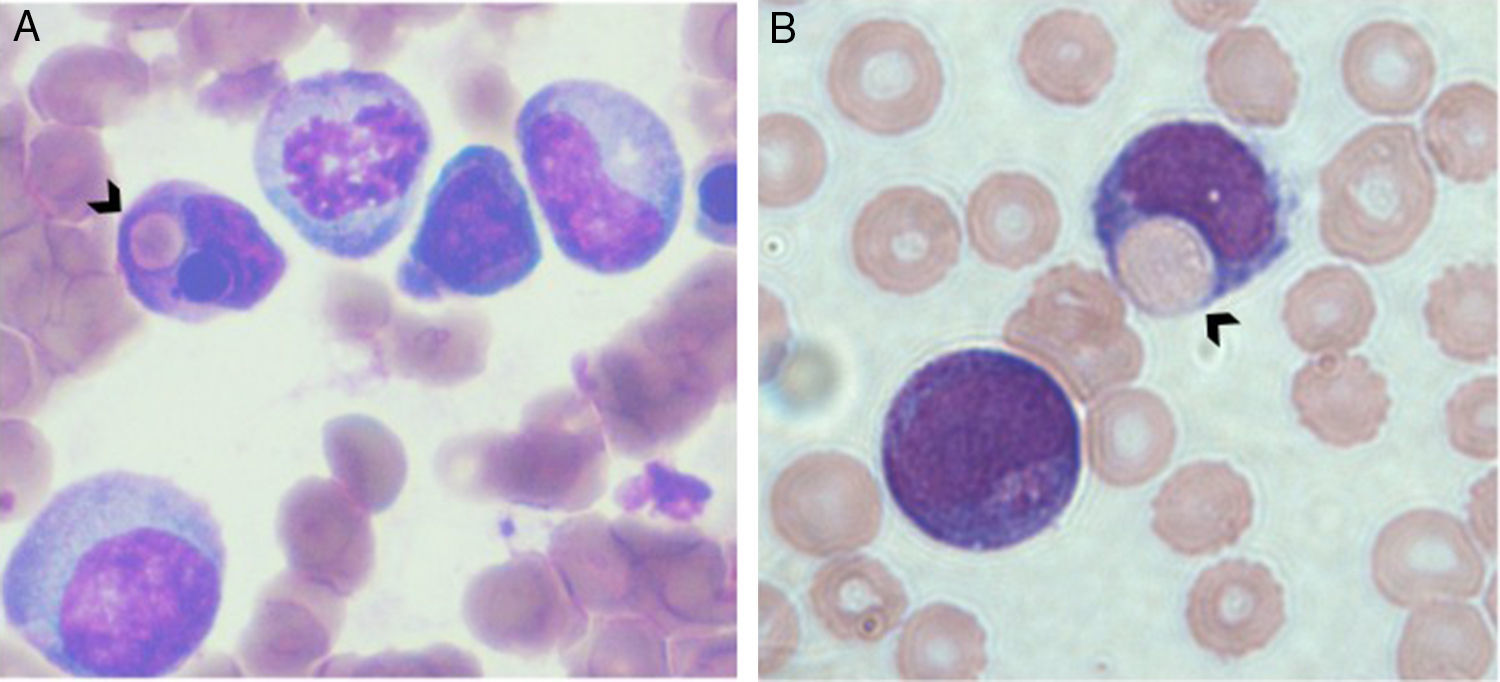
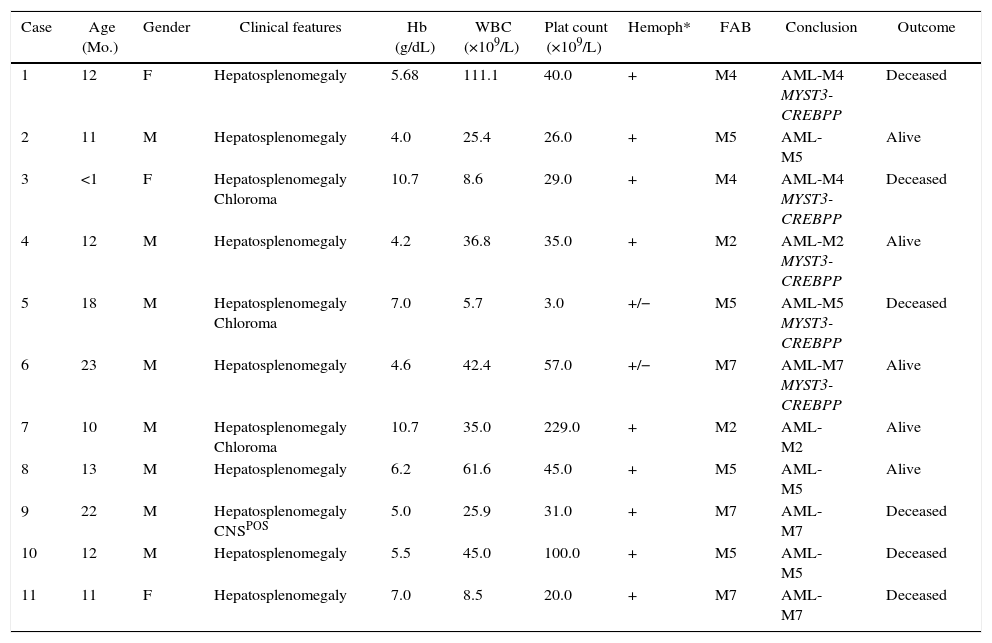
MethodsSubjectsA series of 266 infant AML (i-AML) cases enrolled in the Brazilian Collaborative Study Group of Infant Acute Leukemia (BCSGIAL) from 2003 to 2012 is the reference cohort and subject for the present analysis.9 The selection criteria were infants (≤24months old) with a diagnosis of AML and the presence of hemophagocytosis by leukemic blasts (Figure 1). Additionally, 48 i-AML cases without the hemophagocytic feature in the diagnostic samples were randomly selected to compare with i-AML cases with hemophagocytosis by blast cells.
Hemophagocytosis was defined as the presence of phagocytosis of red cells, lymphocytes and/or platelets only by blast cells. The morphological findings were discussed by physicians (RMB, TCCF, BF, IMQM) and cytologists (EPN, MSPO); clinical and laboratorial data were checked in each case for the consistency of inclusion criteria. Gender, age, white blood cell count (WBC), hemoglobin levels, platelet count, central nervous system (CNS) involvement, chloroma and cutaneous leukemia, FAB classification as well as the presence of hemophagocytosis by leukemic blasts were carefully reviewed. Exclusion criteria included secondary AML, down's syndrome, HLH and/or hemophagocytic syndrome associated with immune disorders and unexplained fever. Frozen samples from bone marrow (BM) aspirates, peripheral blood and smears of i-AML cases were selected for further cytogenetic and molecular studies according to the availability of good biological material.
All children were treated out of clinical trials, but following international AML protocols.
Characterization of leukemia cellsLeukemia classification of AML was based on criteria published by the WHO.7 The diagnosis of AML-M7 was based on the presence of CD41/CD61 and CD42 markers on blast cells identified by immunophenotyping. Karyotypes of BM aspirates were tested before any chemotherapy treatment. Chromosomes were identified and analyzed as recommended by the International System of Human Cytogenetic Nomenclature (ISCN) 2005.10
Reverse transcription polymerase chain reactionTotal RNA from BM mononuclear cells at the time of diagnosis was purified using the TRIzol reagent according to the manufacturer's instructions (Gibco/BRL, Life Technologies, CA, USA). Briefly, 2μg of total RNA was reverse-transcribed using the First-Strand cDNA Synthesis Kit™ (Amersham Pharmacia Biotech Inc., NJ, USA). The integrity of cDNA was examined by amplifying a fragment of the GAPDH gene using previously described primers and cDNA was used as templates in subsequent polymerase chain reaction (PCR) assays. All cases were investigated for the presence of the RUNX1-RUNX1T1, CBFβ-MYH11, BCR-ABL1, MLL-AFF1 and MLL-MLLT3 fusion genes.11–12
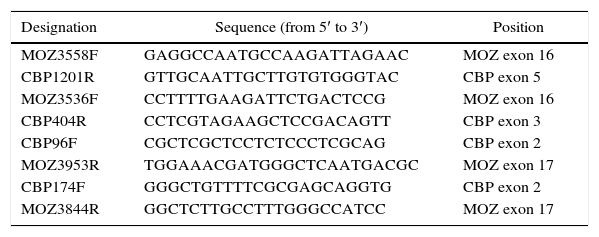
Detection of the MYST3-CREBBP and reverse CREBBP-MYST3 fusion transcripts were conducted as described elsewhere.13 Single PCR reactions to detect MYST3-CREBBP fusion transcripts type I (MYST3 exon 16-CREBBP exon 3) and type II (MYST3 exon 16-CREBBP exon 4), as well as the type I CREBBP-MYST3 fusion transcript (CREBBP exon 2-MYST3 exon 17) were performed using the primers listed in Table 1. A semi-nested reaction, adapted from Schmidt et al., was required to detect type I transcripts.13 Samples from confirmed AML cases with MYST3-CREBBP were added as positive controls for type I transcripts. PCR products for MYST3-CREBBP transcripts (type I-II) were separated by electrophoresis in 1.5% agarose gel and subsequently purified using NucleoSpin Gel and PCR Clean-up kits (Macherey-Nagel, VWR International, Oslo, Norway). Amplicons were mixed with the Big Dye terminator v3.1 Kit (Applied Biosystems) and forward or reverse primers and sequenced in an ABI 3130xl Genetic Analyzer (Applied Biosystems).
Sequences of the primers used for reverse transcription polymerase chain reaction.
| Designation | Sequence (from 5′ to 3′) | Position |
|---|---|---|
| MOZ3558F | GAGGCCAATGCCAAGATTAGAAC | MOZ exon 16 |
| CBP1201R | GTTGCAATTGCTTGTGTGGGTAC | CBP exon 5 |
| MOZ3536F | CCTTTTGAAGATTCTGACTCCG | MOZ exon 16 |
| CBP404R | CCTCGTAGAAGCTCCGACAGTT | CBP exon 3 |
| CBP96F | CGCTCGCTCCTCTCCCTCGCAG | CBP exon 2 |
| MOZ3953R | TGGAAACGATGGGCTCAATGACGC | MOZ exon 17 |
| CBP174F | GGGCTGTTTTCGCGAGCAGGTG | CBP exon 2 |
| MOZ3844R | GGCTCTTGCCTTTGGGCCATCC | MOZ exon 17 |
The identification of somatic mutations in KRAS, FLT3 and c-KIT genes were performed by direct sequencing.14–16
Fluorescence in situ hybridizationFluorescence in situ hybridization (FISH) for the MLL rearrangements was performed at the time of diagnosis with fresh biological material using a commercial LSI MLL Dual Color, Break Apart Rearrangement probe (Cytocell Ltd., Cambridge, UK) according to the manufacturer's instructions. The MYST3-CREBBP FISH was performed in interphase nuclei prepared from frozen viable cells of available cases using bacteria-derived artificial chromosome (BAC). These clones were retrieved from the human genome high resolution BAC re-arrayed clone set available in a web format (http://bacpac.chori.org) and selected according to physical and genetic mapping data reported on Ensembl Browser website (http://www.ensembl.org). DNA was extracted and probes were labeled and hybridized by Blue Genome (Cambridge, UK), with Spectrum Orange or Spectrum Green and validated as a FISH probe set on normal controls. The clones used were RP11-231D20 (chr8:42184655-42188062) and RP11-108L9 (chr8:41832025-41864392) flanking the MYST3 gene (orange) and RP11-387O21 (chr16:3918191-4104380) and RP11-461A8 (chr16:3663996-3693579) flanking the CREBBP gene (green). Procedures were performed according to the manufacturer's instructions. The first step was FISH mapping of clones on normal cells from healthy blood donors in order to confirm their chromosomal location. Cut-off values were calculated as 6±3% of fusion gene signals in 100–300 interphase nuclei.
Ethical considerationsTreatment was approved by local laws and regulations as well as by the Institutional Review Boards of each participating center. Medical informed consent was obtained in accordance with the Declaration of Helsinki. This study was approved by the Research Ethics Committee at the Instituto Nacional de Câncer in Rio de Janeiro, Brazil (CEP/CAEE: 186.688).
ResultsThe clinical-demographic characteristics of the eleven cases that fulfilled the selection criteria are shown in Table 2. All patients presented hepatosplenomegaly and three were reported to have chloroma and one CNS disease. The majority of the patients were male (72.7%) with a median age of 12 months (range: 0–23 months). The WBC count varied from 5.7 to 111.1×109/L with a median of 35.9×109/L; six cases were diagnosed as myelomonocytic leukemia (M4/M5), three cases as AML-M7 and two as AML-M2. Serological tests for viral infections (Epstein–Barr virus, parvovirus B19 and human immunodeficiency virus) and coagulation examinations were within normal ranges. No infections triggering HLH were found in any of the eleven cases.
Demographic and clinical characteristics of selected i-AML cases.
| Case | Age (Mo.) | Gender | Clinical features | Hb (g/dL) | WBC (×109/L) | Plat count (×109/L) | Hemoph* | FAB | Conclusion | Outcome |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 12 | F | Hepatosplenomegaly | 5.68 | 111.1 | 40.0 | + | M4 | AML-M4 MYST3-CREBPP | Deceased |
| 2 | 11 | M | Hepatosplenomegaly | 4.0 | 25.4 | 26.0 | + | M5 | AML-M5 | Alive |
| 3 | <1 | F | Hepatosplenomegaly Chloroma | 10.7 | 8.6 | 29.0 | + | M4 | AML-M4 MYST3-CREBPP | Deceased |
| 4 | 12 | M | Hepatosplenomegaly | 4.2 | 36.8 | 35.0 | + | M2 | AML-M2 MYST3-CREBPP | Alive |
| 5 | 18 | M | Hepatosplenomegaly Chloroma | 7.0 | 5.7 | 3.0 | +/− | M5 | AML-M5 MYST3-CREBPP | Deceased |
| 6 | 23 | M | Hepatosplenomegaly | 4.6 | 42.4 | 57.0 | +/− | M7 | AML-M7 MYST3-CREBPP | Alive |
| 7 | 10 | M | Hepatosplenomegaly Chloroma | 10.7 | 35.0 | 229.0 | + | M2 | AML-M2 | Alive |
| 8 | 13 | M | Hepatosplenomegaly | 6.2 | 61.6 | 45.0 | + | M5 | AML-M5 | Alive |
| 9 | 22 | M | Hepatosplenomegaly CNSPOS | 5.0 | 25.9 | 31.0 | + | M7 | AML-M7 | Deceased |
| 10 | 12 | M | Hepatosplenomegaly | 5.5 | 45.0 | 100.0 | + | M5 | AML-M5 | Deceased |
| 11 | 11 | F | Hepatosplenomegaly | 7.0 | 8.5 | 20.0 | + | M7 | AML-M7 | Deceased |
F: female; M: male; (*), Hemoph: hemophagocytosis (range: 5–27% blasts with phagocytosis); Hb: hemoglobin concentration; i-AML: infant acute myeloid leukemia; Mo: months; FAB: French–American–British classification; NOS: not otherwise specified; Plat: platelet count; WBC: white blood cell count.
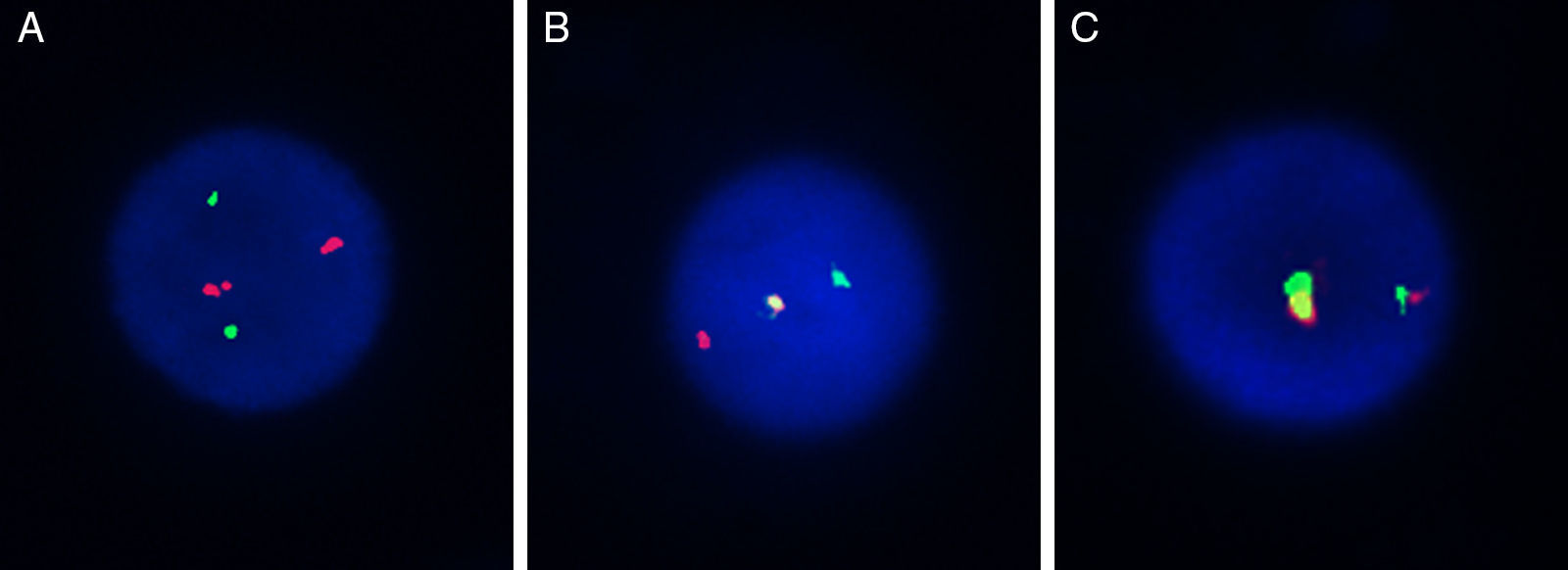
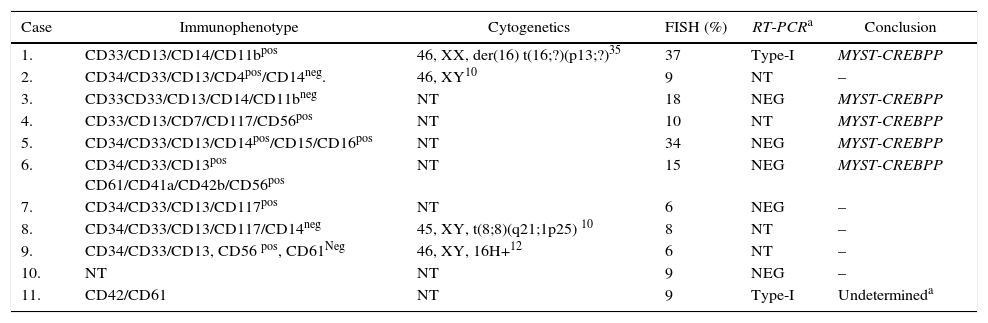
Using the selected BAC clones of the MYST3-CREBBP fusion gene, three types of hybridization patterns were observed (Figure 2). The first, separated signals of the probe combinations RP11-231D20/RP11-108L9 and RP11-387O21/RP11-461A8 on chromosomes 8 and 16 respectively, were observed as four different signals: two red and two green distinct signals consistent with normal chromosomes (Figure 2A). Second, a single fusion pattern was observed (one fusion, one red and one green) considered as a random co-localized signal (Figure 2B). Lastly, a dual fusion signal was found in 15–37% of the interphase nuclei analyzed which was consistent with a breakpoint in MYST3 and CREBBP (Figure 2C).
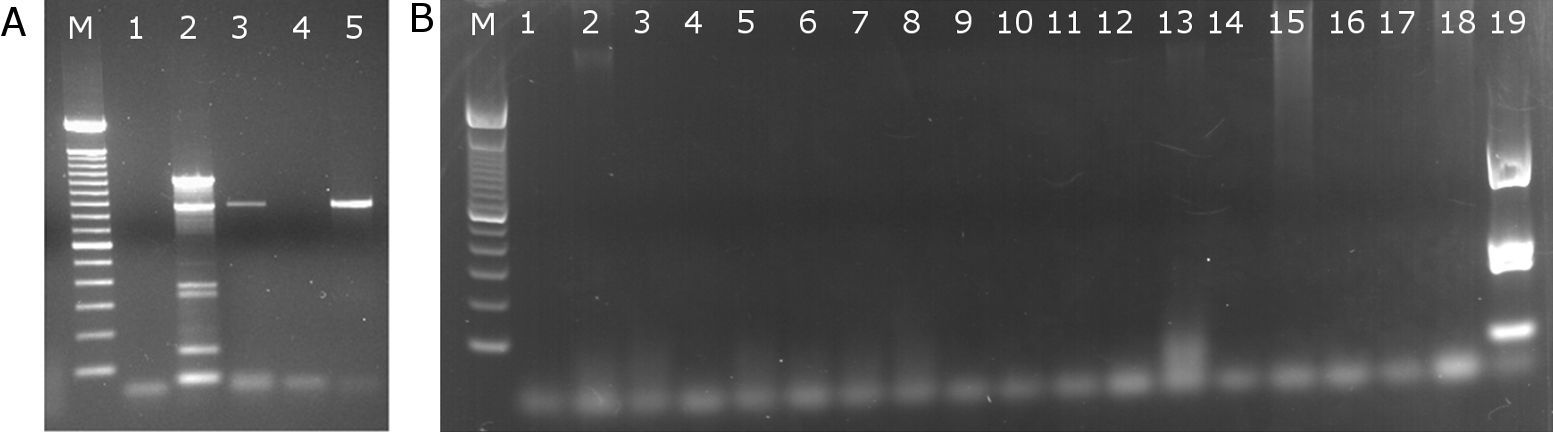
The RT-PCR technique was performed in 55 samples; seven samples were from i-AML with hemophagocytosis (Table 3) and 48 samples from i-AML without hemophagocytosis. The RT-PCR pattern was different to expected (∼1000bp) in one case, with a ∼900bp product observed (#1). In five cases (#3, #5, #6, #7 and #10), RT-PCR was negative for type I and II transcripts. Despite accurate mapping of the translocation breakpoints, attempts to amplify the transcripts, as well as the CREBPP-MYST3 were not successful. In four cases (#2, #4, #8 and #9), RT-PCR was not performed due to lack of suitable biological material. Discrepancies were observed in three cases (#3, #5 and #6) in which dual fusion signals were found in 15–18% and 34% of the interphase nuclei analyzed and RT-PCR results were negative. In all the i-AML cases without hemophagocytosis, the RT-PCR results were negative (Figure 3).
Immunophenotyping, cytogenetic, fluorescence in situ hybridization (FISH) and reverse transcription polymerase chain reaction (RT-PCR) of infant acute myeloid leukemia cases with hemophagocytosis.
| Case | Immunophenotype | Cytogenetics | FISH (%) | RT-PCRa | Conclusion |
|---|---|---|---|---|---|
| 1. | CD33/CD13/CD14/CD11bpos | 46, XX, der(16) t(16;?)(p13;?)35 | 37 | Type-I | MYST-CREBPP |
| 2. | CD34/CD33/CD13/CD4pos/CD14neg. | 46, XY10 | 9 | NT | – |
| 3. | CD33CD33/CD13/CD14/CD11bneg | NT | 18 | NEG | MYST-CREBPP |
| 4. | CD33/CD13/CD7/CD117/CD56pos | NT | 10 | NT | MYST-CREBPP |
| 5. | CD34/CD33/CD13/CD14pos/CD15/CD16pos | NT | 34 | NEG | MYST-CREBPP |
| 6. | CD34/CD33/CD13pos CD61/CD41a/CD42b/CD56pos | NT | 15 | NEG | MYST-CREBPP |
| 7. | CD34/CD33/CD13/CD117pos | NT | 6 | NEG | – |
| 8. | CD34/CD33/CD13/CD117/CD14neg | 45, XY, t(8;8)(q21;1p25) 10 | 8 | NT | – |
| 9. | CD34/CD33/CD13, CD56 pos, CD61Neg | 46, XY, 16H+12 | 6 | NT | – |
| 10. | NT | NT | 9 | NEG | – |
| 11. | CD42/CD61 | NT | 9 | Type-I | Undetermineda |
–: Considered negative; NT: not tested; NEG: Negative.
Agarose gel images of RT-PCR for MYST3-CREBBP fusion genes. Gel A, shows positive reactions for the MYST3-CREBBP fusion gene. Samples 2 (A) and 19 (B) are the positive controls in each reaction. Sample 1 (A and B) is the negative control (H2O only). Samples 3 and 5 (A) are from patients #1 and #11. In gel B, all samples are negative for the MYST3-CREBBP fusion gene; M: standard marker (100 base pairs)
As shown in Table 3, the diagnosis of the MYST3-CREBBP fusion gene was based on the FISH results only or combined with the RT-PCR results. Five cases were diagnosed as AML-MYST3-CREBBP and clinical laboratorial features are summarized: they presented hepatosplenomegaly, skin lesions and/or localized chloroma; hematological tests revealed FAB AML-M2, M4, M5, or M7; the presence of hemophagocytosis by blast cells varied from 5 to 25%. The immunophenotyping profile showed cells positive for the CD34, CD33/CD13/CD14/CD11b/CD14/CD15, CD64, CD56 antigens; in three patients, the blast cells were positive for CD61/CD41a/CD42b/CD56; karyotyping was successful in four cases; one case (#1) revealed a 46, XX, der (16), t(16;?) (p13;?) without identified partners on the short arm of chromosome 16.
None of the eleven AML cases presented with the RUNX1-RUNX1T1, CBFb-MYH11, BCR-ABL1, MLL-AFF1, MLL-MLLT1, KRAS, FLT3 or c-KIT mutations.
The patients received AML treatment according to the BFM AML-2004 protocol and only one out of five patients with the AML MYST3-CREBBP fusion gene is still alive.
DiscussionChromosomal abnormalities in childhood AML are frequent; the MYST3-CREBBP rearrangement, however, is not.1,17 Here, we report for the first time the presence of MYST3-CREBBP rearrangement in five out of eleven (36.4%) AML cases with hemophagocytosis found in a Brazilian i-AML cohort. The morphological observation of hemophagocytosis was an important variable for the selection criteria to investigate the MYST3-CREBBP fusion gene. Clinically, these cases appear to have distinct disease manifestations with skin nodules, CNS involvement and chloroma.1,5,18 As pointed out by Hatano et al., other chromosomal abnormalities in AML, such as t(16;21)(p11;q22), karyotypes involving the 8p11 breakpoint, t(8;19)(p11;q32), complex rearrangements and other chromosomal translocations are associated with the presence of hemophagocytosis by blast cells.19 This supports our data showing an absence of the MYST3-CREBBP fusion gene in four cases in this study. According to the literature, in the majority of patients described with myelomonocytic morphology, the presence of CD56 cellular expression predicts an association with hemophagocytosis and involvement of the leukemia cutis.1,5,19–22
The technique used to identify this chromosomal alteration was FISH because conventional karyotyping was not always available. Failure to obtain mitosis was a pitfall. Multicolor karyotyping technologies such as multicolor-FISH would certainly elucidate such subtle chromosomal rearrangements if mitosis were successfully obtained. Based on hematological signs, we chose to carry out the FISH method followed by RT-PCR as a laboratorial strategy to search for the MYST3-CREBBP fusion gene. The FISH analysis showed a fusion signal above the cutoff value for the specific MYST3 and CREBBP probes on interphase nuclei, suggesting the presence of a MYST3-CREBBP fusion chimera in five cases. This finding indicates a possible generation of MYST3-CREBBP, since both chromosome regions represented by the clones that contain the MYST3 and CREBBP genes appeared co-localized. In one case, RT-PCR for the type I fusion transcript followed by direct sequencing showed a ∼900pb transcript probably related to an unspecific amplification. In the other four cases, no amplicon was observed. Type I (MYST3 exon 16-CREBBP exon 3) is the most common form in adults with MYST3-CREBBP AML.16,23 Low expression or instability of the chimeric transcripts24 and RNA degradation might explain the absence of amplification by RT-PCR.
Recently, Panagopoulos et al. described an AML with hemophagocytosis and with two translocations with breakpoints that suggest other candidate genes different to MYST3 and CREBBP.25 They studied the patients’ leukemic cells not only by karyotyping, FISH and RT-PCR, but also using the modern RNA-seq technique and programs that are specific for fusion genes. Interestingly, the techniques initially failed to detect the biologically important MYST3-CREBBP fusion, although it was manually retrievable from the raw sequencing data, suggesting that additional information about clinical, morphological, and molecular cytogenetic features should be taken into account when searching for newly described crucial fusion genes in typical hematologic malignancies.25
One important point should be discussed is related to AML-MYST3-CREBBP and the differential diagnosis of a hemaphagocytic syndrome such as HLH, which is a severe hyperinflammatory condition with clinical symptoms that include fever, cytopenias, hepatosplenomegaly, and hemophagocytosis.26 However, this hemophagocytosis in BM is morphological in benign macrophages. HLH, when occurring in young children, is associated with inherited genetic defects and diagnostic criteria combine both biological features, including natural killer cell activity and high-soluble interleukin-2-receptor levels.27
AML cases younger than two years old with hemophagocytosis should be investigated for the presence of the MYST3-CREBBP and other chromosomal alterations. In the I-BFM AML study group, more than 50% of the MYST3-CREBBP cases were found in infants and the frequency of congenital cases was significantly higher.5 One of our cases described herein was congenital leukemia. Some authors consider that congenital AML-MYST3-CREBBP may be a self-limiting disease reaching spontaneous remission. A ‘watch-and-wait’ policy should be considered in congenital patients with mild clinical symptoms provided that close long-term monitoring is used.5,28–30 However, cases from our cohort suffered from aggressive disease with dismal outcomes. Interestingly, the genomic landscape of childhood AML-MYST3-CREBBP has a specific signature clustered close to AML with MLL rearrangements.5,31,32 The similarity consists in the characteristic pattern of up-regulation of HOXA9, HOXA10, and cofactor MEIS1 and down-regulation of other homeobox family genes.32 The high frequency of AML-MYST3-CREBBP in infants (≤24 months) and congenital cases support the hypothesis that leukemia occurs during the in utero life and as in the MLL rearrangements model they should be explored for a better understanding of AML leukemogenesis.
Conflict of interestsThe authors declare no conflicts of interest.
The authors thank the physicians’ part of the Brazilian Collaborative Study Group of Infant Acute Leukemia (BCSGIAL) from different Brazilian regions for supporting the project by sending general and clinical data of i-AML.
The authors are grateful to Dr. Tarsis Paiva Vieira for FISH technical support, Bruno de Almeida Lopes for image edition and Dr. Gerhard Fuka for critical and English revision of the manuscript. The authors are also in debt to Dr. Oskar Haas from Anna Kinderspital, Medical University Vienna, Vienna, Austria who kindly provided the AML-MYST3-CREBBP sample to use as positive controls.
MSPO was supported by FAPERJ (#E-26/101.562/2010).