Multiple myeloma (MM) is a prevalent hematological malignancy with high recurrence and no definitive cure. The current study revisits the role of the IGF1/IGF1R axis in MM, introducing a novel inhibitor, NT157. The IGF1/IGF1R pathway is pivotal in MM, influencing cell survival, proliferation, and migration and impacting patient survival outcomes. NT157 targets intracellular proteins such as IRS and STAT proteins and demonstrates antineoplastic potential in hematological malignancies and solid tumors. In the present study, we assessed IGF1R signaling-related gene expression in MM patients and healthy donors, unveiling significant distinctions. MM cell lines displayed varying expression patterns of IGF1R-related proteins. A gene dependence analysis indicated the importance of targeting receptor and intracellular elements over autocrine IGF1. NT157 exhibited inhibitory effects on MM cell viability, clonal growth, cell cycle progression, and survival. Moreover, NT157 reduced IRS2 expression and STAT3, STAT5, and RPS6 activation and modulated oncogenes and tumor suppressors, fostering a tumor-suppressive molecular profile. In summary, our study demonstrates that the IGF1/IGF1R/IRS signaling axis is differentially activated in MM cells and the NT157’s capacity to modulate crucial molecular targets, promoting antiproliferative effects and apoptosis in MM cells. NT157 may offer a multifaceted approach to enhance MM therapy.
Multiple myeloma (MM) stands as the one of most common hematological malignancies originating from B-lymphocytes.1-3 Notwithstanding significant advancements in MM treatments, including the use of proteasome inhibitors, monoclonal antibodies, and immunomodulatory agents, the disease continues to exhibit a high recurrence rate and remains incurable.4,5 Therefore, the identification of new targets or therapeutic strategies is still essential to improve the treatment of this disease.
Here, we revisit the IGF1/IGF1R axis in MM from the perspective of new data and analysis and a novel pharmacological inhibitor in this context. The role of the IGF1/IGF1R pathway has been extensively documented in MM and has been associated with the proliferation, survival, and migration of neoplastic plasma cells, as well as being associated with the prognosis of patients with MM.6-11 In 2013, a novel pharmacological inhibitor of IGF1R/IRS was reported (known as NT157), being the first to act on intracellular targets, the IRS1 and IRS2 proteins.12 Currently, other molecular targets of NT157 have been identified, including STAT3, STAT5, and AXL.13-16 NT157 has been reported as a promising antineoplastic compound in hematological malignancies15,17-19 and solid tumors20-27, but its potential in multiple myeloma is still unknown. Thus, in the present study, the cellular and molecular mechanisms underlying the suppressive effects of NT157 on MM were investigated.
Materials and methodsGene expression data and bioinformaticsIGF1 (median of probes 209540_at, 209541_at, 209542_x_at, and 211577_s_at), IGF1R (median of probes 203627_at and 225330_at), IRS1 (probe 204686_at), and IRS2 (median of probes 209184_s_at and 209185_s_at) mRNA expression data from samples from healthy donors (bone marrow plasma cells [n = 5]) or MM patients (n = 30) were derived from the publicly accessible data portal AmaZonia! database 2008 (http://amazonia.transcriptome.eu).28 The gene expression values were obtained from cDNA microarray experiments by the Affymetrix HUG133 plus 2.0 arrays system and the data were crossed using tumor-specific identification numbers. Gene dependency scores for IGF1, IGF1R, IRS1, and IRS2 derived from high throughput CRISPR-Cas9 screens using twenty MM cell lines (Supplementary Table 1) were obtained from the MyeloDB database (https://project.iith.ac.in/cgntlab/myelodb/).
Cell lines and pharmacological inhibitorsMM.1S, MM.1R, and U266 cells were kindly provided by Prof. Sara Teresinha Olalla Saad (University of Campinas, Campinas, Brazil). RPMI 8226 cells were kindly provided by Prof. Gisele Wally Braga Colleoni (Federal University of São Paulo, São Paulo, Brazil). Cells were cultured in RPMI-1640 supplemented with 10% fetal bovine serum (FBS) plus 1% penicillin/streptomycin and maintained at 5% CO2 and 37ºC. MM cells were used for up to 12 passages after thawing. NT157 was obtained from Sun-Shinechem (Wuhan, China) and prepared as 10 mM stock solutions in dimethyl sulfoxide (DMSO).
Cell viability assayIn total 2 × 104 cells per well were seeded in a 96-well plate in appropriate medium in the presence of vehicle or NT157 (0.8; 1.6; 3.2; 6.4; 12.5; 25 and 50 µM) for 24, 48, and/or 72 h. Next, 10 μl methylthiazoletetrazolium (MTT) solution (5 mg/ml) was added and incubated at 37°C, 5% CO2 for 4 h. The reaction was stopped using 150 μl 0.1N HCl in anhydrous isopropanol. Cell viability was evaluated by measuring the absorbance at 570 nm. IC50 values were calculated using nonlinear regression analysis in GraphPad Prism 8 (GraphPad Software, Inc., San Diego, CA, USA).
Clonogenic assayAutonomous colony formation assays were carried out in semi-solid methylcellulose medium (1 × 103/ml; MethoCult 4230; StemCell Technologies Inc., Vancouver, BC, Canada) in the presence of vehicle or NT157 (1.6, 3.2, 6.4, 12.5, and 25 μM). Colonies were detected after 10-14 days of culture by adding 100 μl (5 mg/ml) MTT reagent and scored using Image J quantification software (U.S. National Institutes of Health, Bethesda, MD, USA).
Cell cycle analysisIn total 1 × 105 cells per well were seeded in 6-well plates supplemented with vehicle or NT157 (0.8, 1.6, 3.2, 6.4, and 12.5 μM), harvested at 24 h, fixed with 70% ethanol and stored at 4°C for at least 4 h. Next, the fixed cells were stained with 20 μg/ml propidium iodide (PI) containing 10 μg/ml RNase A for 30 min at room temperature in a light-protected area. DNA content distribution was acquired using flow cytometry (FACSCalibur; Becton Dickinson, Franklin Lakes, NJ, USA) and analyzed using FlowJo software (Treestar, Inc., San Carlos, CA, USA).
Cell death analysisIn total 4 × 104 cells per well were seeded in a 24-well plate in RPMI-1640 medium with 10% FBS in the presence of vehicle or NT157 (0.8, 1.6, 3.2, 6.4, and 12.5 μM) for 48 h. Next, the cells were washed with ice-cold phosphate-buffered saline (PBS) and resuspended in a binding buffer containing 1 μg/ml propidium iodide (PI) and 1 μg/ml APC-labeled annexin V. All specimens were analyzed by flow cytometry (FACSCalibur; Becton Dickinson) after incubation for 15 min at room temperature in a light-protected area. Ten thousand events were acquired for each sample.
Western blot analysisTotal protein extraction was performed from MM.1S and MM.1R cells treated with vehicle or NT157 (3.2 and 6.4 µM) for 24h using a buffer containing 100 mM Tris (pH 7.6), 1% Triton X-100, 150 mM NaCl, 2 mM PMSF, 10 mM Na3VO4, 100 mM NaF, 10 mM Na4P2O7, and 4 mM EDTA. Equal amounts of protein were used from total extracts followed by SDS-PAGE and a Western blot analysis with the indicated antibodies, as previously described.16 Western blot analysis was performed using a SuperSignalTM West Dura Extended Duration substrate system (Thermo Fisher Scientific, San Jose, CA, USA) and a G: BOX Chemi XX6 gel document system (Syngene, Cambridge, UK). Antibodies against phospho(p)-IGF1RTyr1135 (#3918), IGF1R (#3027), IRS1 (#3407), IRS2 (#3089), p-STAT3Tyr705, (#9131) STAT3 (#4905), p-STAT5Tyr694 (#9359), STAT5 (#25656), p-RPS6Ser235/236 (#4858), RPS6 (#2217), p-ERK1/2Thr202/204 (#9101), ERK1/2 (#9102), PARP1 (#9542), γH2AX (#9718), and α-tubulin (#2144) were obtained from Cell Signaling Technology (Danvers, MA, USA).
Quantitative PCRTotal RNA from MM.1S and MM.1R cells treated with vehicle or NT157 (6.4 µM) for 24h was obtained using the TRIzol reagent (Thermo Fisher Scientific). cDNA was synthesized from 1 µg of RNA using a High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). The quantitative PCR (qPCR) was performed using a QuantStudio 3 Real-Time PCR System in conjunction with a SybrGreen System using specific primers for NT157-related genes (Supplementary Table 2).15-18HPRT1 and ACTB were used as reference genes. Relative quantification values were calculated using the 2−ΔΔCT equation.29 A negative “No Template Control” was included for each primer pair. Data were visualized using the multiple experiment viewer (MeV) 4.9.0 software.30
Statistical analysisStatistical analyses were performed using GraphPad Prism 8 (GraphPad Software, Inc.). For comparisons, Mann-Whitney, Student t-test or ANOVA test and Bonferroni post-test were used. Correlation analysis was performed using the Spearman test and RStudio software (version 1.4.1717, RStudio, PBC) and the Corrplot plugin. A p-value < 0.05 was considered statistically significant.
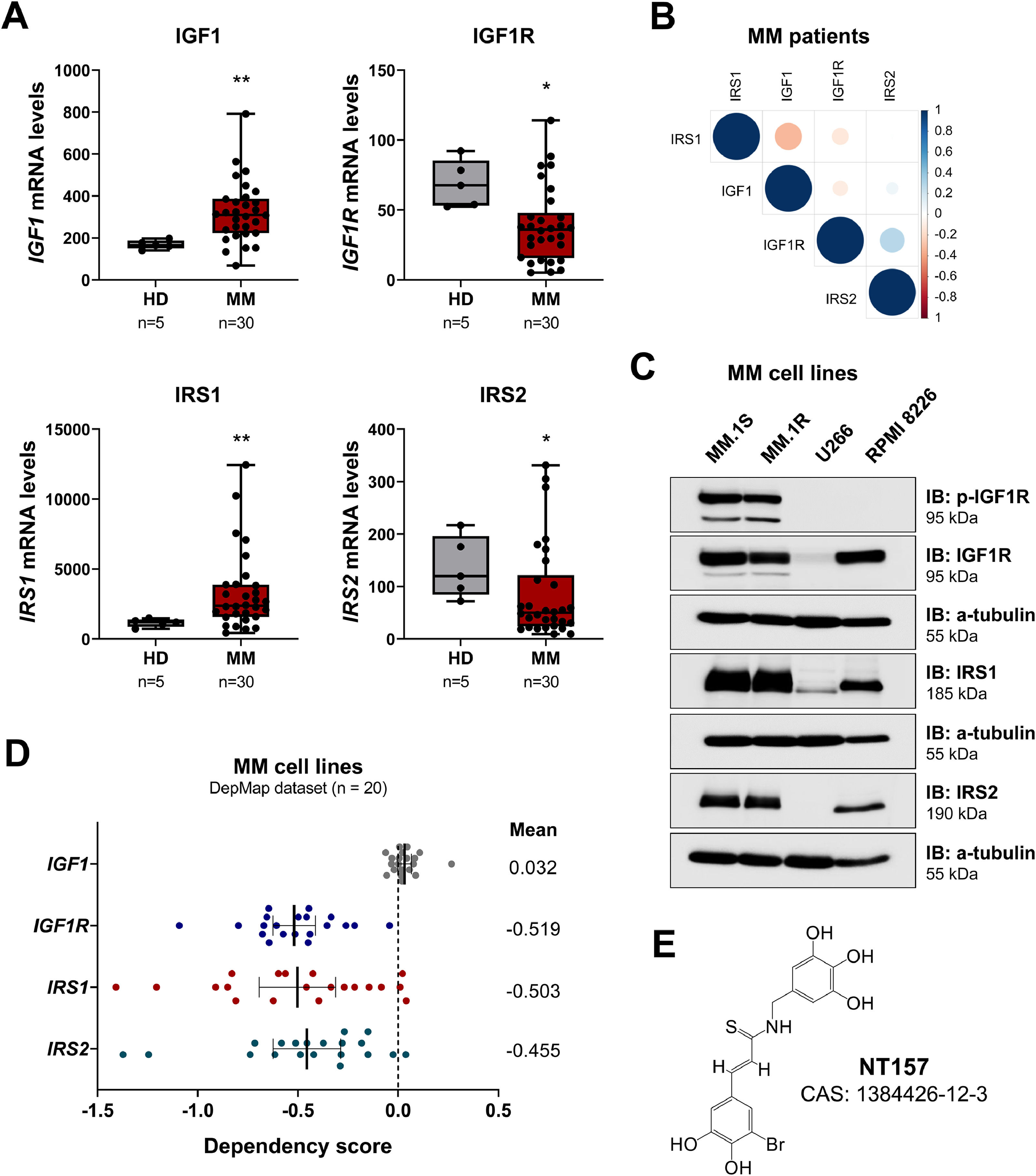
ResultsIGF1R signaling-related genes are differentially expressed and present different levels of dependence in multiple myelomaFirstly, the levels of genes related to IGF1R-mediated signaling were evaluated in healthy donors and patients with multiple myeloma. IGF1 and IRS1 mRNA expression were significantly increased, while IGF1R and IRS2 mRNA expression were significantly decreased in MM patients (all p < 0.05; Figure 1A). A positive correlation was observed between the expression of IGF1R and IRS2 (r = 0.38, p = 0.03; Figure 1B) in MM patients. MM cell lines showed a heterogeneous expression of elements related to IGF1R-mediated signaling. MM.1S and MM.1R cells showed the highest levels of p-IGF1R, IGFR, IRS1, and IRS2, while U266 cells showed low levels of IGF1R and IRS1 and did not show detectable expression of IRS2 and p-IGF1R. RPMI 8226 cells showed high levels of IGF1R, IRS1, and IRS2 but not p-IGF1R (Figure 1C).
Differential expression of IGF1R signaling-related genes in multiple myeloma patients. (A) IGF1, IGF1R, IRS1, and IRS2 mRNA levels in samples from healthy donors and multiple myeloma (MM) patients. The y-axis represents gene expression data obtained from the AmaZonia! database 2008, which were measured using Affymetrix HGU133 plus 2.0 arrays. The data sets were cross-referenced using tumor-specific identification numbers. The numbers of subjects for each group are indicated. The p-values are also indicated; Mann–Whitney U test. (B) Correlogram of IGF1R signaling-related genes in MM patients. The correlation analysis was performed using the Spearman test and RStudio software and the Corrplot plugin. The sizes of the circle indicate the r values and the color indicates the direction of the correlation (blue: positive correlation, red: negative correlation). (C) Western blot analysis p-IGF1R, IGF1R, IRS1, and IRS2 in the total extract from MM cell lines. The membranes were re-probed for the detection of α-tubulin and developed using a SuperSignal™ West Dura Extended Duration Substrate system and a G:BOX Chemi XX6 gel doc system. (D) Gene dependency scores for IGF1, IGF1R, IRS1, and IRS2 of 20 MM cell lines were obtained from the MyeloDB database. For gene effect, a score less than -0.5 represents depletion in most cell lines, while less than -1 represents strong killing. (E) Representation of the NT157 chemical structure. CAS, Chemical Abstracts Service.
The dependence score was 0.032 for IGF1, -0.519 for IGF1R, -0.503 for IRS1, and -0.455 for IRS2, indicating that the receptor and intracellular targets of the pathway are more relevant as targets than the autocrine production of IGF1, which could be obtained via paracrine pathways (Figure 1D). Therefore, NT157, a multitarget inhibitor, was selected for functional studies (Figure 1E). Among the targets of NT157 are the proteins IRS1/2 and STAT3/5.12,13,15
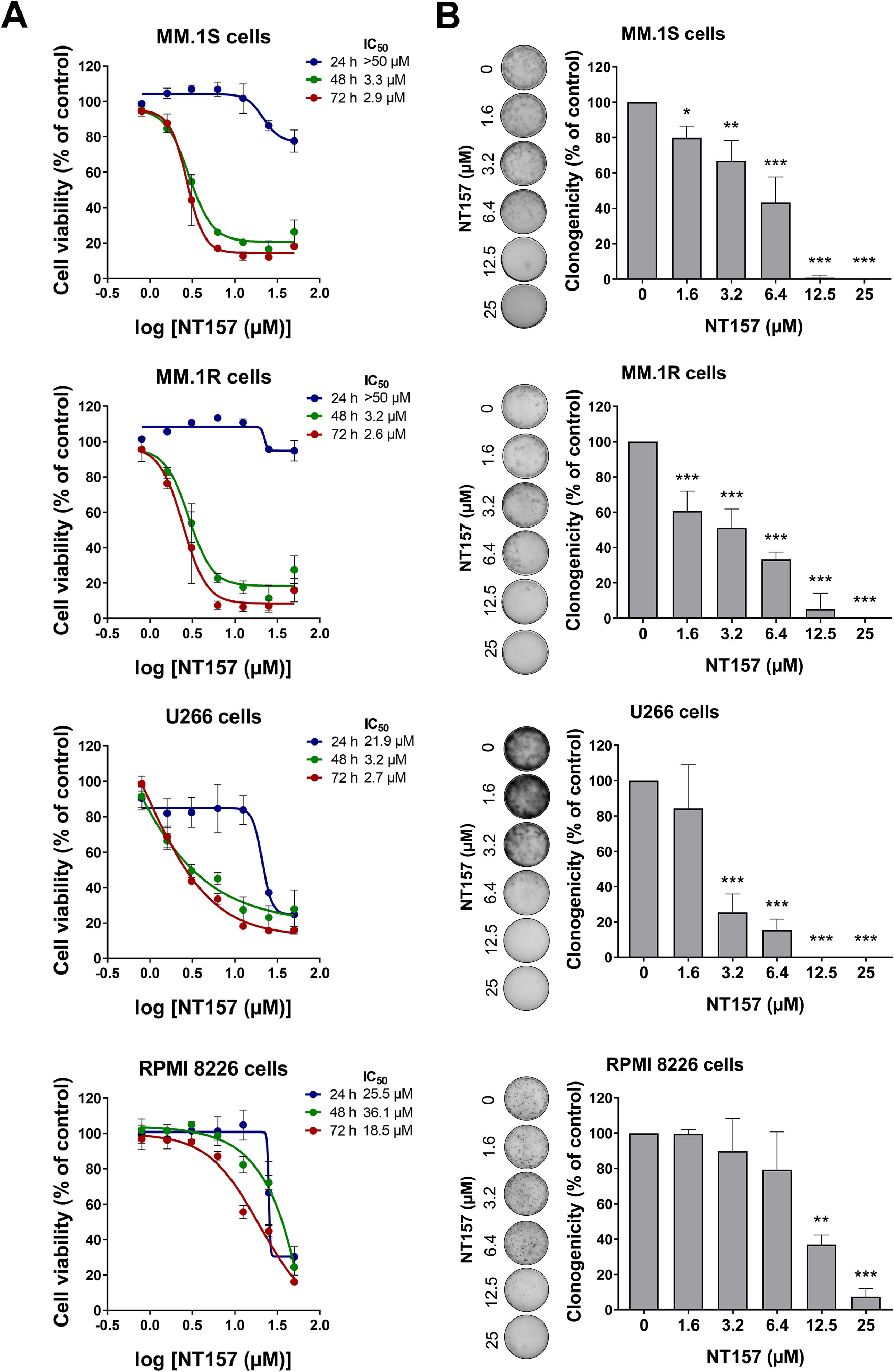
Multiple myeloma cell viability and autonomous clonal growth are suppressed by NT157Next, we investigated the effects of NT157 on cell viability and autonomous clonal growth of MM cells. NT157 reduced the dose- and time-dependent manner cell viability of all MM cells evaluated, being more potent and efficient in MM.1S, MM.1R, and U266 cells (Figure 2A). IC50 values ranged from 2.9 to >50 µM for MM.1S cells, from 2.6 to >50 µM for MM.1R cells, from 2.7 to 21.9 µM for U266 cells, and from 18.5 to 36.1 µM for RMPI 8226 cells. NT157 also strongly reduced autonomous clonal growth in all MM cell lines (all p < 0.05; Figure 2B), which suggests that NT157 had a suppressive effect on the ability of MM cells to grow and multiply independently.
NT157 reduces cell viability and autonomous clonal growth of multiple myeloma cells. (A) Dose- and time-response cytotoxicity was analyzed by methylthiazoletetrazolium (MTT) assay for multiple myeloma (MM) cells treated with graded concentrations of NT157 (ranged from 0.8 to 50 µM) for 24, 48, and 72 h. Values are expressed as the percentage of viable cells for each condition relative to vehicle-treated controls. Results are shown as the mean ± SD of at least three independent experiments. (B) Colonies containing viable cells were detected by adding an MTT reagent after 10-14 days of culturing the cells in the presence of vehicle or NT157 (ranged from 1.6 to 25 µM). Colony images are shown for one experiment and bar graphs show the mean ± SD of at least three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001; ANOVA test and Bonferroni post-test.
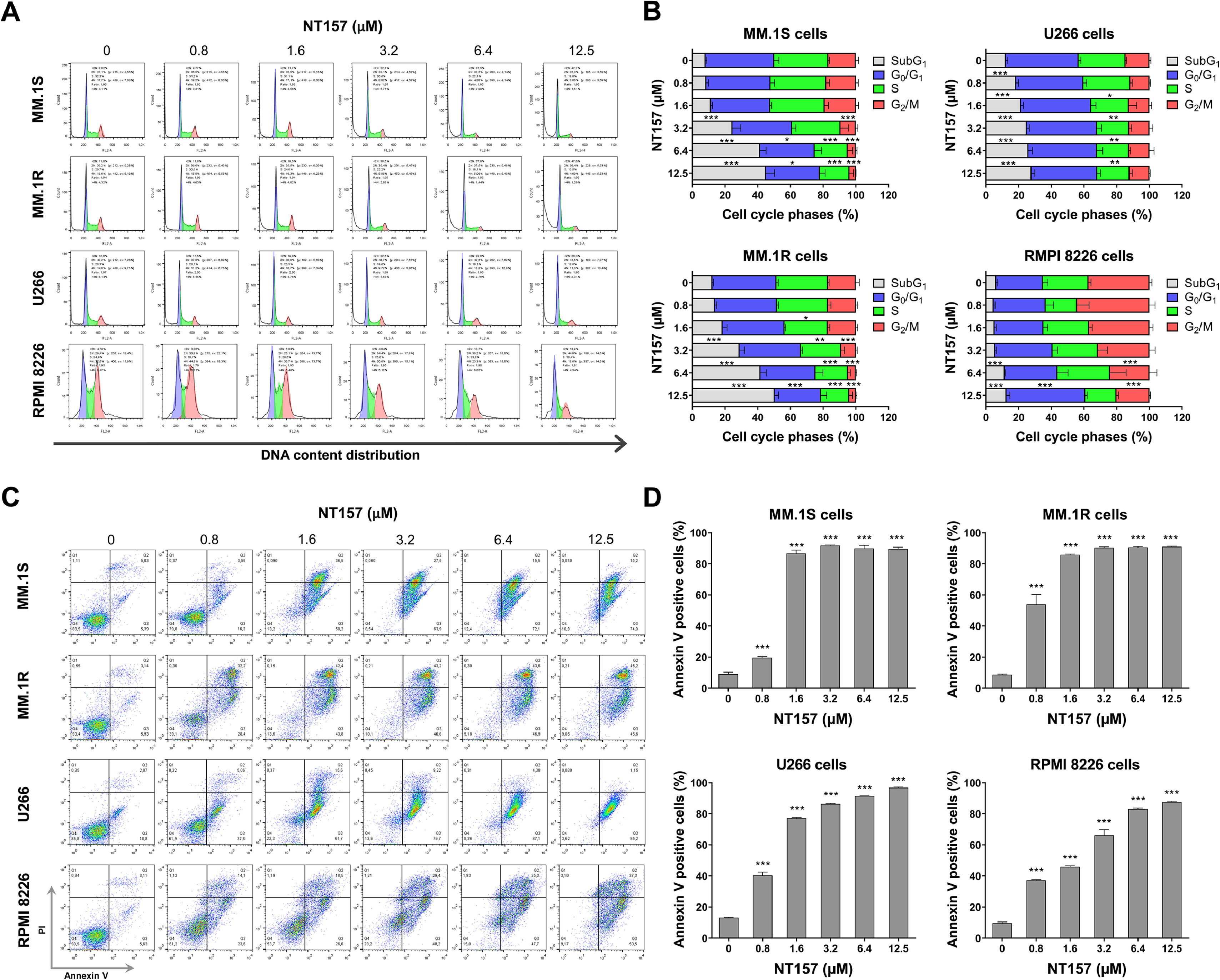
In an effort to gain insight into the cellular mechanisms induced by NT157 that lead to the reduction in cell viability, we conducted additional experiments. The analysis of cell cycle progression revealed that NT157 had a significant effect on the increase of cell population in subG1 and reduced the number of cells in the proliferative phases (S and G2/M phases) in the MM cell models studied (all p < 0.05, Figure 3A). The increase in the subG1 population suggests that NT157 is inducing cell cycle arrest, particularly in cells that have damaged DNA or are undergoing programmed cell death. Complementing these results, the annexin V externalization analyses indicated that NT157 was a potent inducer of apoptosis at concentrations as low as 0.8 µM after 48 h of exposure (all p < 0.05, Figure 3B). This further confirms the compound's ability to trigger cell death pathways in MM cells.
NT157 triggers cell cycle arrest and cell death in multiple myeloma cells. (A) Cell cycle progression was determined by flow cytometry in multiple myeloma (MM) cells treated with the indicated concentrations of NT157 for 24 h. A representative histogram for each condition is illustrated. (B) Bar graphs represent the mean ± SD of the percent of cells in subG1, G0/G1, S, and G2/M phase upon vehicle or NT157 exposure (ranging from 0.8 to 12.5 µM) for 24 h and represent at least three independent experiments. The p values and cell lines are indicated in the graphs. *p < 0.05, **p < 0.01, ***p < 0.001; ANOVA test and Bonferroni post-test. (C) Cell death was detected by flow cytometry in MM cells treated with vehicle or NT157 (ranging from 0.8 to 12.5 µM) for 48 h using an annexin V/PI staining method. Representative dot plots are shown for each condition; the upper and lower right quadrants (Q2 plus Q3) cumulatively contain the cell death population (annexin V+ cells). (D) Bar graphs represent the mean ± SD of at least three independent experiments. The p values and cell lines are indicated in the graphs. ***p < 0.001; ANOVA test and Bonferroni post-test.
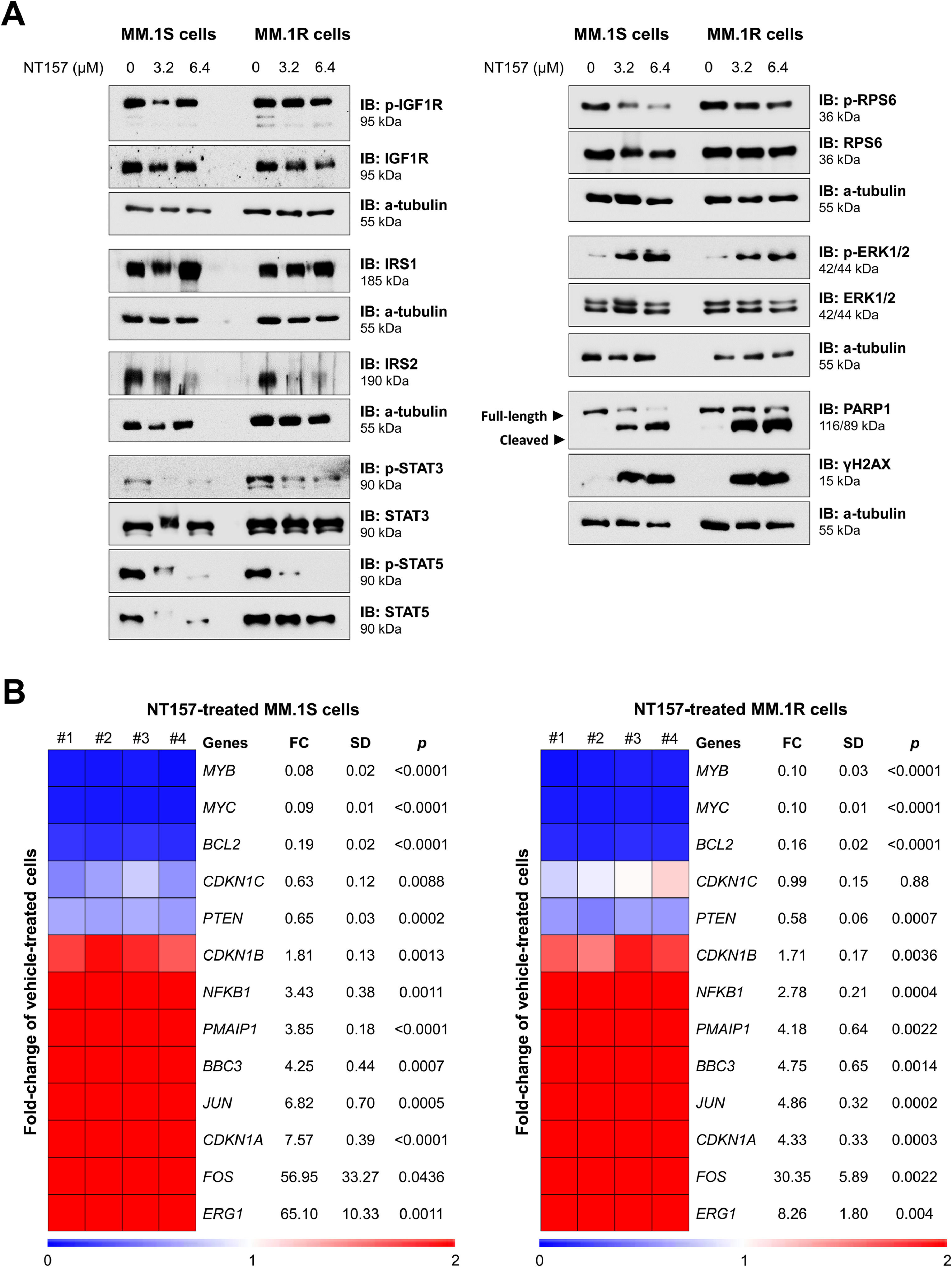
The effects of NT157 were investigated on molecular targets previously associated with the compound and which participate in cell proliferation and survival. NT157 reduced the expression of IRS2 and the activation of STAT3, STAT5, and RPS6. The compound increases the activation of ERK1/2, a mechanism previously associated with cellular stress and degradation of IRS proteins.12 Markers of apoptosis (cleaved PARP1) and DNA damage (γH2AX) were also strongly induced by the compound (Figure 4A). Finally, NT157 reduced the expression of important oncogenes such as MYB, MYC, and BCL2 and increased the expression of tumor suppressors associated with cellular stress (ERG1, FOS, and JUN), cell cycle arrest (CDKN1A and CDKN1B), and apoptosis in response to DNA damage (BBC3 and PMAIP1) (all p < 0.05, Figure 4B). Overall, these results suggest that NT157 exerts its anti-proliferative and pro-apoptotic effects on multiple myeloma cells by modulating a network of key molecular targets involved in cell survival and growth, and promoting DNA damage and apoptosis.
NT157 favors a tumor-suppressive molecular profile in multiple myeloma cells. (A) Western blot analysis phospho(p)-IGF1R, IGF1R, IRS1, IRS2, p-STAT3, STAT3, p-STAT5, STAT5, p-RPS6, RPS6, p-ERK1/2, ERK1/2, PARP1 (total and cleaved), and γH2AX in the total extract from MM.1S and MM.1R cells treated with vehicle or NT157 (6.4 µM) for 24 h. The membranes were re-probed for the detection of α-tubulin and developed using a SuperSignal™ West Dura Extended Duration Substrate system and a G:BOX Chemi XX6 gel doc system. (B) Heatmap of the gene expression in MM.1S and MM.1R cells treated with vehicle or NT157 (3.2 and 6.4 μM). The data are represented as the fold-change of vehicle-treated cells, and downregulated and upregulated genes are shown by blue and red colors, respectively. Genes, fold-change (FC), standard deviation (SD) and p values (Student t test) are indicated.
In the present study, we report the expression of genes associated with IGF1R signaling in MM patients, as well as the effects of NT157, a pharmacological inhibitor of the IGF1R/IRS axis, in cellular models of MM. In our study, IGF1 and IRS1 mRNA levels were increased, while IGF1R and IRS2 mRNA levels were reduced in MM patients. The importance of IGF1 has been widely reported to be relevant for the proliferation, survival, and migration of MM cells.6-10 The autocrine production of IGF1 has been identified as key to cell survival under serum-free culture conditions in MM cells.7 Furthermore, serum IGF1 levels and IGF1R expression were associated with worse clinical outcomes in MM patients.7,11 Although the role of IGF1R and its ligand, IGF1, are well described in the literature, the functions of the adapter proteins IRS1 and IRS2 are no less known. It has been demonstrated that constitutive IRS2 tyrosine phosphorylation and PI3K recruitment mediated by IGF1R are key players in the development of a broad spectrum of murine plasma cell tumors.31 IGF1-induced proliferation and antiapoptotic effects in human multiple myeloma cell lines through the activation of the PI3K/AKT and MAPK pathways, dependent on IRS1. This effect was observed even in cell lines that were not responsive to IL6, underscoring the significant role of the IGF1R/IRS1 axis in driving the development and progression of this disease.32
Interestingly, in the present study, gene dependence studies have indicated that inhibition of autocrine IGF1 production was less relevant than inhibition of the receptor, IGF1R, and the intracellular targets, IRS1 and IRS2. In fact, several studies have shown that multiple myeloma cells may obtain IGF1 from other sources, stimulating cells in the bone marrow microenvironment such as mesenchymal stem cells, osteoblasts, and endothelial cells.33 These findings drew our attention to NT157, a pharmacological inhibitor initially described as an IGF1R/IRS inhibitor12, but which has currently been associated with the inhibition of multiple targets of interest in oncology such as STAT3, STAT5 and AXL.13-16 NT157 exhibits antineoplastic effects in a wide variety of leukemias15,17-19 and solid tumors.20-27
From a molecular perspective, NT157 exhibited several notable effects in MM cells. It reduced the expression of IRS2 and suppressed the activation of STAT3, STAT3, and RPS6. While previous studies have documented crosstalk between IRS2 and STAT proteins in hematopoietic cells and hematologic neoplasms34, NT157 targets both of them through distinct mechanisms. IRS2 is degraded via proteasomal degradation, mediated by ERK1/2 activation.12 In parallel, STAT3 and STAT5 undergo dephosphorylation through the activation of protein phosphatases.13,15 RPS6, a ribosomal protein downstream target of the PI3K/AKT/mTOR pathway, regulates cell growth and survival by promoting protein synthesis.35
Furthermore, NT157 downregulated the expression of pivotal oncogenes, including MYC, MYB, and BCL2. MYC dysregulation is well-documented in MM, as it often participates in primary IgH translocations36 and is considered a potential therapeutic target for the disease.37 Similarly, BCL2 has been identified as dysregulated and a promising target in MM.38,39 Within the BCL2 family, NT157 increased the expression of pro-apoptotic members such as BBC3 (also known as PUMA) and PMAIP1 (also known as NOXA), which may contribute to the induction of cell death mediated by compound.40-42
Our study presents opportunities for targeting the IGF1R/IRS axis in MM using NT157, elucidating the primary molecular changes involved in the loss of cell viability triggered by the drug. However, it is important to acknowledge certain limitations. The available data for both healthy donors and MM patients are limited, preventing a comprehensive clinical-laboratory association analysis. Additionally, it is crucial to highlight that the experimental data are derived from cell lines. Future studies using patient cells in ex vivo assays and murine models are warranted to further validate our findings.
In summary, the IGF1/IGF1R/IRS signaling axis is differentially activated in MM cells, and the gene dependence analysis suggests that the presence of the receptor and intracellular adapter proteins are more important than the autocrine IGF1, probably that this may be obtained from paracrine sources. Pharmacological inhibition of IGF1R/IRS1 showed marked antineoplastic effects in cellular models of MM. The molecular analysis highlights NT157′s propensity to support a tumor-suppressive molecular profile, emphasizing the need for future investigations into its potential therapeutic benefits.
G.N.Q., K.L., and L.B.L.M. received a fellowship from the São Paulo Research Foundation (FAPESP; grants #2023/01331-2, #2020/12842-0, and #2022/03316-8). This study was supported by grant #2021/11606-3 from the FAPESP. This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Brasil (CAPES), Finance Code 001.