Acute myeloid leukemia (AML) is a disease characterized by clonal proliferation and accumulation of myeloid progenitor cells in the bone marrow (BM), inhibition of cell differentiation, increased proliferative index and defective apoptosis.1 AML secondary to myelodysplastic syndrome (MDS–sAML) is characterized by diverse cytogenetic and molecular changes including del(5q), as well as changes in the RNA splicing pathway, TET2, EZH2, FLT3, NRAS, NPM1, RUNX1, DNMT3a, IDH1, IDH2, TET2, TP53 genes, etc.2 Established prognostic factors in AML include age, the cytogenetic and molecular profile and history of hematologic disorders, such as MDS.1 About 40% of elderly AML patients were previously diagnosed with MDS, which is usually refractory to chemotherapy.1 We present a case of a sAML, which showed a single and previously not described abnormality, a chromosomal translocation between chromosomes 8 and 13 t(8;13) identified by conventional cytogenetics, and several altered genes detected by DNA microarray assay, suggesting that the t(8;13) rearranged region results in altered gene expression patterns.
Case reportThe patient was a 72-year-old female admitted to the Instituto Nacional de Cancer (INCA), Rio de Janeiro, Brazil. She presented with a history of increasing fatigue and dyspnea. Past medical history included arterial hypertension, coronary artery disease, myalgia and pneumonia. The laboratory results at diagnosis revealed: white blood cell count 36.9×109/L with 41% monocytoid blasts, hematocrit 23%, hemoglobin 7.6g/dL, platelets 131.0×109/L and lactate dehydrogenase 806IU/L. A bone marrow aspirate showed 34% myeloid and monocytoid blasts (FAB M4) with dysplastic features in erythroid and myeloid cells. Flow cytometry immunophenotyping identified CD33+, CD14+, CD11b+, CD13+, CD38−, HLA-DR−, CD15+, MPO−, CD117+, TDT−, cCD79a−, CD2−, and CD7− cells. Reverse transcription-polymerase chain reaction (RT-PCR) for RUNX1/RUNX1T1, BCR/ABL1 and MYH11/CBFB, showed no fusion transcripts. G-banding revealed an abnormal karyotype and the gene expression profile was investigated for altered genes in the rearranged t(8;13) region.
This study was approved by the institutional review board of INCA (number 110/06). The study was conducted in accordance with the Helsinki Declaration as revised in 2008.
MethodsCytogenetic analysis was performed on unstimulated BM cells for 24h according to standard protocols with the karyotype being described according to the International System for Human Cytogenetic Nomenclature (ISCN, 2013).3 The gene expression profile was investigated using an Affymetrix GeneChip Human Gene 1.0 ST Array (Affymetrix Inc., Santa Clara, CA, USA) following the manufacturer's instructions to identify differentially expressed genes in the rearranged t(8;13) region. Array data were extracted and processed with the open software packages from the Bioconductor Project (www.bioconductor.org). In brief, the data was normalized with Robust Multi-Array Average expression measure. Subsequently, a non-specific filter was applied with the genefilter package4 in order to remove Affymetrix probes and genes that exhibited low variance across samples; differentially expressed genes were selected using the linear models for microarray data package and summarized at log2>2. RNA extracted from sAML peripheral blood was compared to normal peripheral blood using a pool of six healthy donors. Patient's relative gene expression levels were compared to the control sample level and lists of ‘up-regulated’ and ‘down-regulated’ genes were generated by the program.
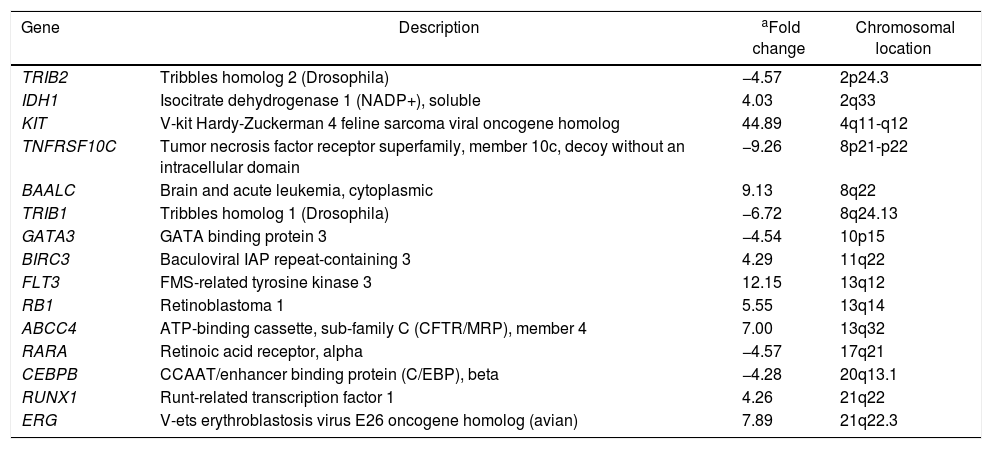
ResultsG-banding revealed an abnormal karyotype with a translocation involving chromosome 8 and 13, resulting in the previously undescribed karyotype 46,XY,t(8;13)(q22;q11)[4]/46,XX[28] (Figure 1) according to the catalogs of cancer cytogenetics.5 The microarray analysis labeled 874 as differentially expressed in over 28,000 genes. The gene expression levels (fold change), shown in Table 1, demonstrated down-regulated genes involved in myeloid development or apoptosis, including CEBPB, RARA, GATA3, TRIB1, TRIB2 and TNFRSF10C (TRAIL-R3). In addition, up-regulated genes related to proliferation, differentiation and drug resistance, such as KIT, IDH1, ERG, BIRC3 and ABCC4 were observed by microarray analysis. The gene RUNX1 (AML1) required for the generation of definitive hematopoietic stem cells during embryogenesis was up-regulated. The brain and acute leukemia cytoplasmic (BAALC) gene, located in the 8q22 region, and the FLT3 and RB1 genes, located in the 13q12 and 13q14 regions, respectively, appear to be intimately linked to the t(8;13). These genes were up-regulated suggesting that they could be altered due to the rearrangement of this region. Another gene that may have been inactivated due to this rearrangement is the TRIB1 gene, located in the 8q24.13 region, which is a potent negative regulator of MAPK signaling.
Up-regulated and down-regulated genes in t(8;13) acute myeloid leukemia secondary to myelodysplastic syndrome.
| Gene | Description | aFold change | Chromosomal location |
|---|---|---|---|
| TRIB2 | Tribbles homolog 2 (Drosophila) | −4.57 | 2p24.3 |
| IDH1 | Isocitrate dehydrogenase 1 (NADP+), soluble | 4.03 | 2q33 |
| KIT | V-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | 44.89 | 4q11-q12 |
| TNFRSF10C | Tumor necrosis factor receptor superfamily, member 10c, decoy without an intracellular domain | −9.26 | 8p21-p22 |
| BAALC | Brain and acute leukemia, cytoplasmic | 9.13 | 8q22 |
| TRIB1 | Tribbles homolog 1 (Drosophila) | −6.72 | 8q24.13 |
| GATA3 | GATA binding protein 3 | −4.54 | 10p15 |
| BIRC3 | Baculoviral IAP repeat-containing 3 | 4.29 | 11q22 |
| FLT3 | FMS-related tyrosine kinase 3 | 12.15 | 13q12 |
| RB1 | Retinoblastoma 1 | 5.55 | 13q14 |
| ABCC4 | ATP-binding cassette, sub-family C (CFTR/MRP), member 4 | 7.00 | 13q32 |
| RARA | Retinoic acid receptor, alpha | −4.57 | 17q21 |
| CEBPB | CCAAT/enhancer binding protein (C/EBP), beta | −4.28 | 20q13.1 |
| RUNX1 | Runt-related transcription factor 1 | 4.26 | 21q22 |
| ERG | V-ets erythroblastosis virus E26 oncogene homolog (avian) | 7.89 | 21q22.3 |
sAML develops in approximately 40% of patients with MDS and the clinical discrimination between AML and MDS is based on cytomorphological analysis, since patients with MDS have dysplastic hematopoiesis and a myeloblast count of less than 20%, whereas those with a myeloblast count of 20% or more have AML.6 sAML has clinical and biological heterogeneity linked to chromosome aberrations or molecular changes with the association between them suggesting that those mechanisms are significantly involved in leukemogenesis.1 This case report shows evidence that t(8;13)(q22;q11) could be involved in the pathogenesis and severity of AML. The translocation t(8;13) with breakpoints at (8q22) and (13q11) has neither been reported nor described for possible altered genes. The gene expression profile was performed to determine the specific signature in cells from this patient and to try to clarify a new possible molecular pathway involved in disease evolution. Of the 874 genes differentially expressed, we focused mainly on genes related to AML pathogenesis, prognosis and response to standard therapy, and the genes located in the altered chromosome region. Important genes such as CEBPB, which plays a pivotal role in proliferation and differentiation, including suppression of myeloid leukemogenesis,7RARA, which is required for optimal myelomonocytic differentiation7 and GATA3 associated with erythropoiesis8 were down-regulated as was the tumor suppressor gene TRIB2 and TNFRSF10C (TRAIL-R3) which is important in initiating apoptosis.9,10 Genes involved with anti-apoptotic functions and resistance to drug therapy such as BIRC3 and ABCC4,11,12 as well as genes that participate in the inhibition of differentiation or induction of proliferation such as, KIT, IDH1 and ERG were up-regulated.13–15 Another gene that was up-regulated is the RUNX1 gene, which acts as a regulator of the expression of various genes specific to hematopoiesis, and plays an important role in myeloid differentiation. RUNX1 amplification is associated with increased risk of relapse and worse overall outcome in AML.16 The BAALC gene, located in the 8q22.3 region, is a novel molecular marker indicating an unfavorable outcome in AML with normal cytogenetics.17 Its high expression may act as an adverse prognostic factor through prompting cell proliferation and inhibiting apoptosis in leukemia cells such as in AML, acute lymphoblastic leukemia (ALL), and chronic myelogenous leukemia in blast crisis patients.17 High levels of the FLT3-wild-type receptor may promote constitutive activation of this receptor in malignant cells and is associated with a worse prognosis and high risk of relapse in pediatric AML patients.18 The RB1 gene, located in the 13q14 region, is known as a tumor suppressor; abnormalities affecting the RB1 pathway are not always related to gene expression. These abnormalities are usually associated with a lack of the pRB protein or the expression of a mutant pRB, and inappropriate phosphorylation of pRB, leading to deregulated G1-S transition, that frequently occurs in malignant disorders.19 There are alternative mechanisms that down-regulate pRB protein levels that include reduced translation of the RB1 mRNA, or reduced half-life of the mRNA and proteins having a role in leukemogenesis.19 The Tribbles gene (TRIB-1) is a potent negative regulator of MAPK pathways that influence apoptosis, differentiation and cell-cycle progression. It is known as a tumor suppressor and is usually down-regulated in AML.9 The expression of the BAALC, FLT3 and RB1 genes in this specific region appears relevant for the pathogenesis of AML as they could be regulated together in one regulatory module. Such combined expression suggests a probable higher functional relevance when compared to the expression of each gene independently.
ConclusionsThere are no reports in the literature of this type of rearrangement in patients with sAML, nor of the altered genes we found in this region, contributing to the pathogenesis of AML. Although there is not enough material to study protein fusion by next-generation sequencing, we found important up- and down-regulated genes involved in hematopoiesis in this case of t(8;13). A better understanding of the molecular processes affected by the fusion of these genes involved in sAML may shed light on the role of this translocation in leukemogenesis, disease progression and its prognostic effects.
ContributionsARP and RCM were involved in designing the study and the literature search on the subject; RCM was involved in gathering the clinical data; LOC was involved in performing the cytogenetic analysis; FCCF and MBP were involved in performing gene expression analysis; ARP, FCCF, LOC; MBP and RCM were all involved in writing and editing the manuscript.
Conflicts of interestThe authors declare no conflicts of interest.
The authors thank to Marcos Antonio M. Scheiner for his invaluable comments. This study was supported by INCT, CNPq and FAPERJ.


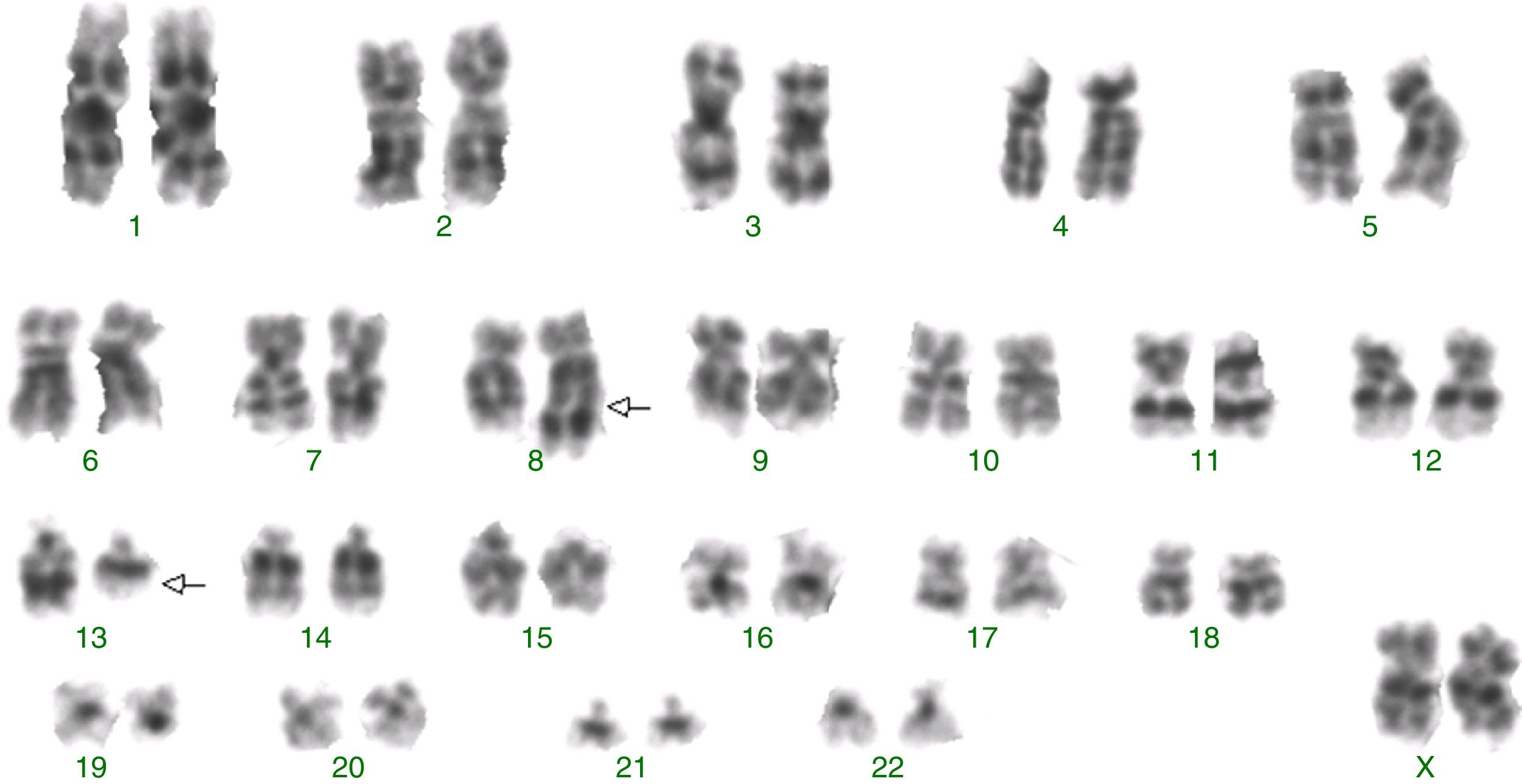
![Abnormal karyotype identified by G-banding with a translocation involving chromosome 8 and 13: 46,XY,t(8;13)(q22;q11)[4]/46,XX[28]. Abnormal karyotype identified by G-banding with a translocation involving chromosome 8 and 13: 46,XY,t(8;13)(q22;q11)[4]/46,XX[28].](https://static.elsevier.es/multimedia/15168484/0000003900000004/v2_201911281025/S1516848417300944/v2_201911281025/en/main.assets/thumbnail/gr1.jpeg?xkr=ue/ImdikoIMrsJoerZ+w93OM6WmS6o9DeZl+SVh74uo=)


