Acute myeloid leukemia (AML) disease entities are classified by the World Health Organization (WHO) based on significant cytogenetic and molecular genetic findings.1,2 The category of acute myeloid leukemia not otherwise specified (AML-NOS) includes cases with ≥20% myeloblasts in the peripheral blood (PB) or bone marrow (BM) in the absence of recurrent genetic abnormalities, myelodysplasia-related changes or previous cytotoxic therapy. Cases of AML with dysplasia in ≥50% of the cells in two or more myeloid cell lineages or cases preceded by a well-documented history of myelodysplastic syndromes (MDS) or MDS/myeloproliferative neoplasm are defined as AML with myelodysplasia-related changes (AML-MRC).1,2 AML-MRC develops in approximately one-third of MDS and generally has a poor prognosis.3,4 Cytogenetics is a mandatory tool in the diagnosis of AML with chromosome abnormalities being present in approximately half of the adult AML cases; chromosome abnormalities have an important prognostic value.5
Metaphase cytogenetics has proven to be an extremely valuable clinical tool in the management of hematological malignancies. This methodology can detect balanced chromosomal changes, including translocation or inversion, and unbalanced chromosomal changes, including trisomy, duplication, and deletion. The success of metaphase cytogenetics requires cellular proliferation and chromosome spreads; the sensitivity and the resolution depend on the proportion of clonal cells in the tested sample and on the size of the lesion, respectively.6 Single nucleotide polymorphism array (SNP-A), also referred to as chromosomal microarray, has been applied as a high-resolution whole genome scanning tool to detect unbalanced chromosomal changes. The major advantage of SNP-A over metaphase cytogenetic analysis is its ability to detect hidden chromosomal defects, including submicroscopic (cryptic) aberrations and the distinction of individual genotypes to detect copy number-neutral loss of heterozygosity (CN-LOH), also defined as uniparental disomy.7
The Philadelphia (Ph) chromosome results from a balanced translocation t(9;22) (q34;q11.2) that leads to the formation of the fusion protein BCR-ABL1 with constitutive tyrosine kinase activity. Three different breakpoint cluster regions in the BCR gene (M-bcr, m-bcr, and μ-bcr) are reported: 8.5kb hybrid mRNA (b2a2 or b3a2) encodes the 210kDa protein (p210), 7.5kb hybrid mRNA (e1a2) encodes the 190kDa protein (p190) and 9kb hybrid mRNA (c3a2) encodes the 230kDa protein (p230).8 The t(9;22)(q34;q11.2) is found in 90–95% of cases of chronic myeloid leukemia and around 20% of acute lymphoblastic leukemia. In contrast, the Ph chromosome is very rare in AML, accounting for approximately 1–2% of the cases.9 Neuendorff et al.10 recently reviewed case reports and case series of AML with BCR-ABL1 in the literature since 1975, as well as cases from their own institution. The authors reported that among the 126 confirmed cases of AML with BCR-ABL1, 38% were defined by WHO 2008 as AML-NOS, 32.5% as AML-MRC, and 16.7% as core binding factor leukemia.10
Recently, the WHO 2016 classification included a new provisional category for de novo AML with BCR-ABL1, recognizing this rare diagnostic entity that may benefit from tyrosine kinase inhibitor therapy.11 The patients are stratified as intermediate II risk according to European LeukemiaNet and in the poor-risk group according to the National Comprehensive Cancer Network.5,12
Herein, we present two unusual cases of AML with BCR-ABL1. In accordance with WHO 2008 definitions, Case 1 was diagnosed as AML-MRC and presented with complex karyotype and the e1a2 BCR-ABL1 gene fusion. Case 2 was diagnosed as de novo AML-NOS and presented a near-tetraploidy karyotype and the e1a2 BCR-ABL1 gene fusion. Both cases were investigated by convectional cytogenetics and molecular approaches.
MethodsPatientsPatients described in this report were followed up at the Hospital das Clínicas of the Universidade de São Paulo in Ribeirão Preto, São Paulo, Brazil. The Ethics Committee of the institution approved the study, and written informed consent was obtained. The algorithm proposed by Neuendorff et al.10 was used to exclude the diagnosis of chronic myeloid leukemia blast crisis.
Metaphase cytogeneticsMetaphase cytogenetics was performed on BM aspirate using standard methods and the karyotype was described according to the International System for Human Cytogenetic Nomenclature (ISCN) 2013.13
Single nucleotide polymorphism arrayFor the SNP-A, genomic DNA was extracted from BM according to the manufacturer's instructions (QIAGEN DNA Kit, Valencia, CA, USA). SNP-A was performed using Affymetrix Genome-Wide Human SNP Cytoscan HD (Affymetrix, Santa Clara, CA, USA). The files were analyzed using Chromosome Analysis Suite (ChAS) software. Regions of copy number variants (CNVs) larger than 1Mb and CN-LOH larger than 10Mb were denoted as true aberrations. In order to detect the somatic origin copy number alterations distinguished from constitutional polymorphic CNVs, the lesions identified using SNP-A were compared with the Database of Genomic Variants (DGV; http://projects.tcag.ca/variation). Aberrations that were identified by SNP-A were described according to ISCN 2013.13
Reverse transcriptase-polymerase chain reaction to detect BCR-ABL1Total RNA (1μg) was obtained from the patient's peripheral blood cells and submitted to reverse transcription polymerase chain reaction (RT-PCR) using the High Capacity cDNA Reverse Transcription Kit (Life Technologies, Carlsbad, CA, USA). The first PCR was performed on a volume of 25μL containing: 1× reaction buffer, 1.5mM MgCl2, 200μM dNTPs, 10pmol of each primer (Mbcr1: 5′-GAAGTGTTTCAGAAGCTTCTC-3′; 2oabl1: 5′-TGATTAAGCCTAAGACCC GGA-3′; mbcr1: 5′-CCATCGTGGGCGTCCGCA-3′), 0.5U Taq DNA polymerase (Life Technologies) and 1.5μL cDNA. The first PCR conditions were an initial phase of 2min at 94°C, then 25 cycles at 94°C for 30s, 51°C for 40s, 72°C for 1min, followed by a final extension step at 72°C for 7min. The second PCR was performed on a volume of 25μL containing: 1× reaction buffer, 1.5mM MgCl2, 200μM dNTPs, 10pmol of each primer (Mbcr2: 5′-TGGAGCTGCAGATGCTGACCAACTC-3′; mbcr2: 5′-AGATCTGGCCCAACG ATGGCGAGGGC-3′; 2iabl2: 5′-ATCTCCAGTGGCCAGAAAATCATAC-3′), 0.5U Taq DNA polymerase (Life Technologies) and 1μL of PCR products from the first reaction. Second PCR conditions were an initial phase of 2min at 94°C, then 35 cycles at 94°C for 30s, 60°C for 30s, 72°C for 1min, followed by a final extension step at 72°C for 7min. The PCR products were analyzed on 2% agarose gel stained with ethidium bromide.
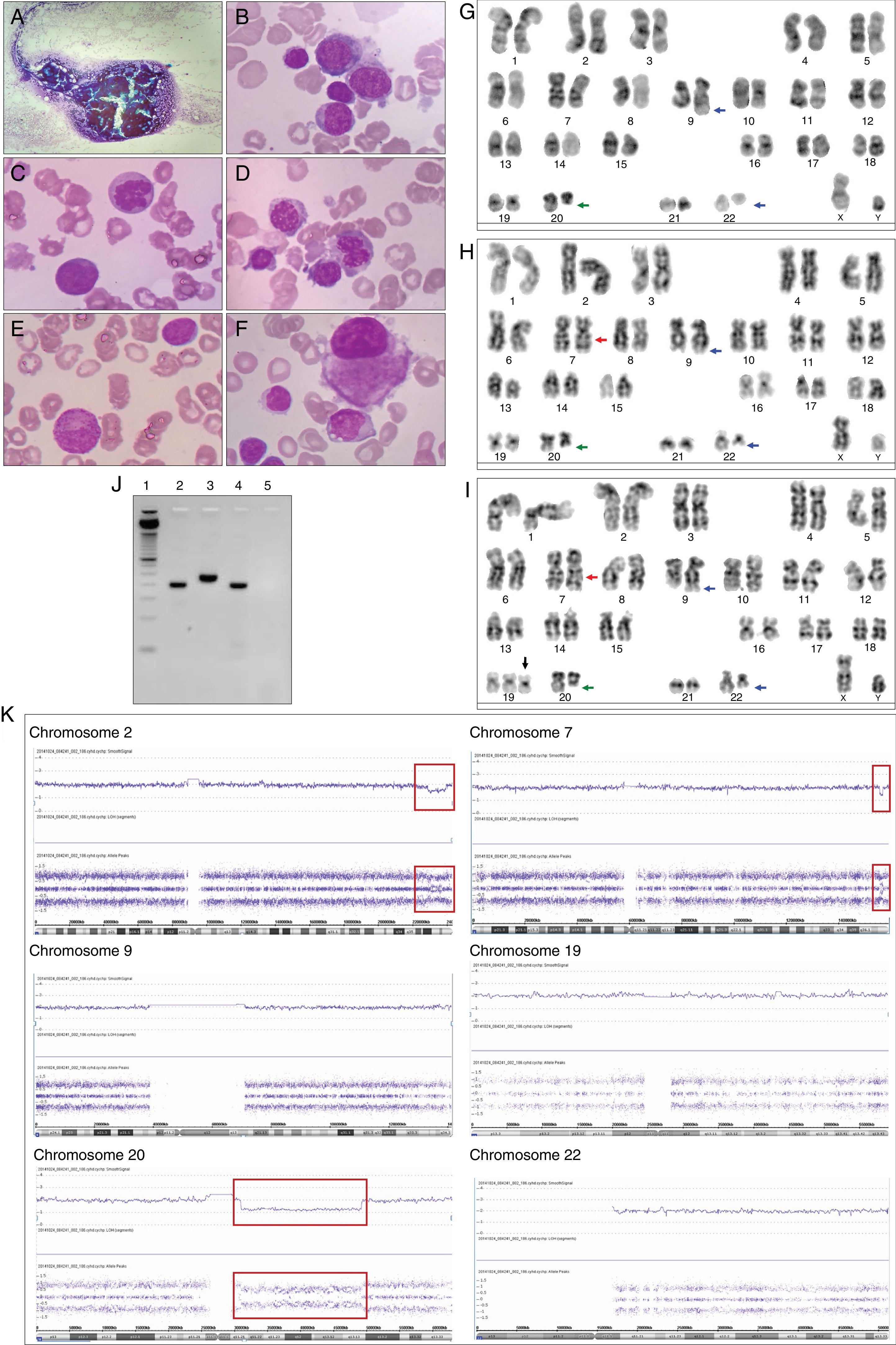
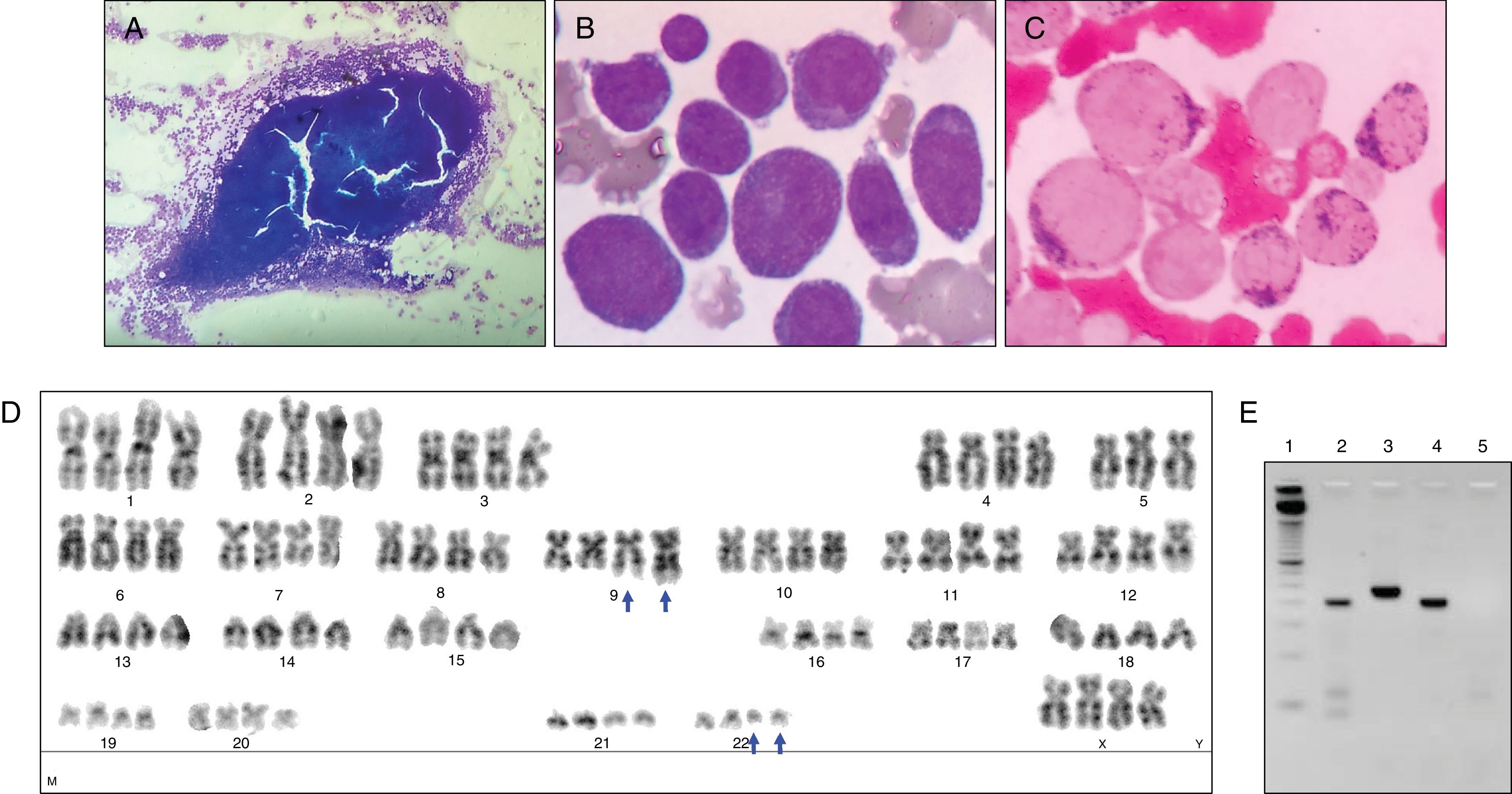
Case 1An 82-year-old male with systemic hypertension, without other relevant medical history and good performance status (Eastern cooperative oncology group: 0) sought medical attention complaining of weight loss, asthenia, and petechiae during the previous 45 days. Physical examination revealed pallor, petechiae and absence of hepatomegaly and splenomegaly. The complete blood count showed pancytopenia (hemoglobin 7.2g/dL, white blood cell count 2.5×109/L, neutrophil count 0.3×109/L, lymphocyte count 2.2×109/L, and platelet count 8×109/L). BM aspiration revealed 20% myeloblasts, and dysplastic alterations in 70% of erythroid and megakaryocytic lineages (Figure 1A–F). The immunophenotypic analysis showed that the blasts were positive for human leukocyte antigens – (HLA)-DR, CD13, CD33, CD34, CD117, CD7, CD133, and CD56 and negative for CD19, CD15, CD11c, CD42a, CD22, CD2, CD11b, CD64, CD14, and CD36. The patient was started on subcutaneous low dose cytarabine immediately after the diagnosis of AML. Metaphase cytogenetics analysis revealed 46,XY,t(9;22)(q34;q11),del(20)(q11)[2]/46,idem,inv(7)(q22q36)[6]/47,idem,inv(7)(q22q36),+19[12] (Figure 1G–I). An RT-PCR assay for BCR-ABL1 confirmed the presence of t(9;22) and revealed a p190 BCR-ABL1 isoform (e1a2 BCR-ABL1) (Figure 1J). The SNP-A identified the loss of 2q36 not identified by metaphase cytogenetics: arr[hg19] 2q36.3q37.3(229,385,664-239,698,667)x1, and confirmed the loss of 20q11: arr[hg19] 20q11.21q13.13(30,955,339-49,197,975)x1. As expected, the SNP-A did not detect the balanced translocation t(9;22), neither the inversion of 7q. SNP-A allowed for the identification of the loss of 7q36 not identified by metaphase cytogenetics: arr[hg19] 7q36.3(155,607,058-156,777,458)x1. However, SNP-A failed to identify the trisomy of chromosome 19, probably explained by the limited sensitivity of the test for lesions present in less than 30% of the uncultured cells (Figure 1K). The patient was classified as AML with BCR-ABL1 according to WHO 2016 and as AML-MRC according to WHO 2008. The patient presented persistent pancytopenia and increased BM blasts following two cycles of subcutaneous low dose cytarabine and was started on imatinib. No hematologic response was observed after two months of imatinib and the patient subsequently died of infectious complications.
Bone marrow morphological, cytogenetics and molecular analysis of Case 1. (A) Hypercellular bone marrow fragment. The bone marrow was stained with Wright-Giemsa stain and the images show: (B) myeloblasts, (C) dyserythropoiesis with karyorrhexis, (D) dyserythropoiesis with binucleate erythroid precursor and nuclei/cytoplasm asynchronous maturation, (E) myeloid precursor with hypogranulation cytoplasm and (F) dysplastic small megakaryocytes with monolobed nuclei. The G-banded karyotype revealed 46,XY,t(9;22)(q34;q11),del(20)(q11)[2]/46,idem,inv(7)(q22q36)[6]/47,idem,inv(7)(q22q36),+19[12]; illustrative metaphase (G–I). The t(9;22)(q34;q11) is indicated with blue arrows, the del(20)(q11) is indicated with a green arrow, the inv(7)(q22q36) is indicated with a red arrow and the +19 is indicated with a black arrow. (J) Polymerase chain reaction for b2a2/b3a2 and e1a2 BCR-ABL1 transcripts – 1: ladder 50 base pairs (Life Technologies); 2: Case 1; 3: positive control for b2a2/b3a2 BCR-ABL1 transcript (p210); 4: positive control for e1a2 BCR-ABL1 transcript (p190); 5: negative control for b2a2/b3a2 and e1a2 BCR-ABL1 transcripts. (K) Single nucleotide polymorphism array (SNP-A)-based karyotyping using Affymetrix Genome-Wide Human SNP Cytoscan HD. Signals at the top of each panel represent copy number status. The lower panel represents genotyping calls or the frequency of A and B alleles. Deletions are illustrated by a decrease in the copy number and a change in genotyping; interstitial deletion in chromosome 2, 7 and 20 are highlighted with a red box. The del(7)(q36) was not identified by metaphase cytogenetics. Normal copy number and three genotypes were observed in chromosome 19 by SNP-A. Chromosomes 9 and 22 presented normal copy number and genotypes by SNP-A.
A 28-year-old woman presented with leukocytosis (white blood cell count 47.8×109/L, myeloblasts 66%, metamyelocytes 1%, neutrophil band 1%, segmented neutrophils 4%, lymphocytes 28%), anemia (hemoglobin 6.9g/dL), and thrombocytopenia (33×109/L). The patient had been suffering from weakness and fatigue associated with febrile episodes for one month. There was no past medical history of any hematological disorder. Physical examination revealed pallor and absence of hepatomegaly and splenomegaly. BM aspiration showed myeloblasts comprising 88% of all cells (Figure 2A–C). The immunophenotypic analysis showed that the blasts were positive for CD34, CD117, CD33, CD13, HLA-DR, CD38, CD11b, CD4, CD133, CD15, CD11c and CD64 and negative for CD7, CD56, CD 19, CD41 and NG2. The patient was diagnosed with de novo AML-NOS and classified as acute myelomonocytic leukemia according to WHO 2008 or AML with BCR-ABL1 according to WHO 2016. The cytogenetic analysis showed 92<4n>,XXXX,−4,−5,+6,−7,−11,+12,−13,−14,+15,−17,−21,t(9;22)(q34;q11)x2[cp20] (Figure 2D) and a RT-PCR assay for BCR-ABL1 revealed a p190 isoform (e1a2 BCR-ABL1) (Figure 2E). The patient received two cycles of induction therapy with daunorubicin and cytarabine, associated with intrathecal chemotherapy (methotrexate, cytarabine and dexamethasone). At the second cycle of induction, the patient was started on dasatinib. The patient achieved complete hematological remission and received matched related myeloablative allogeneic hematopoietic stem cell transplantation. At the time of this report, twenty months after diagnosis, she remains in complete remission.
Bone marrow morphological, cytogenetics and molecular analysis of Case 2. (A) Hypercellular bone marrow fragment. (B) The bone marrow was stained by Wright-Giemsa stain and a high frequency of myeloblasts was observed. (C) The bone marrow was stained by myeloperoxidase stain. (D) Illustrative metaphase: 91<4n>,XXXX,−5,t(9;22)(q34;q11)x2. The t(9;22)(q34;q11.2) is indicated with blue arrows. (E) Polymerase chain reaction assay for b2a2/b3a2 and e1a2 BCR-ABL1 transcripts – 1: ladder 50 base pairs (Life Technologies); 2: Case #2; 3: positive control for b2a2/b3a2 BCR-ABL1 transcript (p210); 4: positive control for e1a2 BCR-ABL1 transcript (p190); 5: negative control for b2a2/b3a2 and e1a2 BCR-ABL1 transcripts.
Herein, we described the characteristics of two cases of AML with BCR-ABL1. Although very rare, the presence of the Ph chromosome has been reported in some MDS cases at diagnosis or at the time of disease progression, and both BCR-ABL1 variants, p210 and p190, have been previously described.9,14,15 In a cohort containing 148 patients with the Ph chromosome, Keung et al.9 reported only two cases of de novo AML (1%) and three cases of MDS (2%). Fukunaga et al.14 reported a case of MDS that presented an abrupt evolution to AML-MRC and the acquisition of the Ph chromosome (p210 BCR-ABL1). In that report, the patient had hematological response to nilotinib, which was lost with the acquisition of the BCR-ABL1 Y253H mutation. Subsequently, the patient had transient hematological response to dasatinib, but response was lost with the acquisition of the BCR-ABL1 T315I and E255K mutations.
Case 1 illustrates a case of AML with BCR-ABL1 with complex karyotype including del(20)(q11) and inv(7)(q22q36) detected by metaphase cytogenetics and loss of 7q36 identified by SNP-A. Partial or whole deletions of the long arm of chromosome 7 are clearly associated with poor prognosis. Previous studies of myeloid malignancies demonstrated that the 7q22 chromosome band is involved in most cases with 7q deletions in MDS and AML.16,17 Deletion of the long arm of chromosome 20 is a common recurring chromosomal abnormality associated with myeloid malignancies.18
In AML, the near-tetraploidy karyotype is a rare cytogenetic alteration and its prognostic relevance is not well defined.19 Yamaguchi et al.15 reported a case of AML with BCR-ABL1 who presented metaphase cytogenetics with 92,XXYY,t(9;22)(q34;q11)x2[20] and RT-PCR positivity for p190 BCR-ABL1; the patient achieved complete remission after the first cycle of induction chemotherapy and was consolidated with allogeneic hematopoietic stem cell transplantation.15
In conclusion, cytogenetic abnormalities identified by karyotyping remain an important prognostic factor in AML. The report of these rare cases of AML with the presence of e2a1 BCR-ABL1 and future analyses for these alterations in AML patients are important to define the local incidence, to improve diagnosis and classification, and to better assess the impact of these molecular alterations on the outcomes of AML patients.
FundingThis work received financial support from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP).
Conflicts of interestThe authors declare no conflicts of interest.
The authors would like to thank Andy Cumming for the English review, and Amélia Góes and Claudia Helena Magnani for their valuable technical assistance with RT-PCR for BCR-ABL1 detection.


![Bone marrow morphological, cytogenetics and molecular analysis of Case 1. (A) Hypercellular bone marrow fragment. The bone marrow was stained with Wright-Giemsa stain and the images show: (B) myeloblasts, (C) dyserythropoiesis with karyorrhexis, (D) dyserythropoiesis with binucleate erythroid precursor and nuclei/cytoplasm asynchronous maturation, (E) myeloid precursor with hypogranulation cytoplasm and (F) dysplastic small megakaryocytes with monolobed nuclei. The G-banded karyotype revealed 46,XY,t(9;22)(q34;q11),del(20)(q11)[2]/46,idem,inv(7)(q22q36)[6]/47,idem,inv(7)(q22q36),+19[12]; illustrative metaphase (G–I). The t(9;22)(q34;q11) is indicated with blue arrows, the del(20)(q11) is indicated with a green arrow, the inv(7)(q22q36) is indicated with a red arrow and the +19 is indicated with a black arrow. (J) Polymerase chain reaction for b2a2/b3a2 and e1a2 BCR-ABL1 transcripts – 1: ladder 50 base pairs (Life Technologies); 2: Case 1; 3: positive control for b2a2/b3a2 BCR-ABL1 transcript (p210); 4: positive control for e1a2 BCR-ABL1 transcript (p190); 5: negative control for b2a2/b3a2 and e1a2 BCR-ABL1 transcripts. (K) Single nucleotide polymorphism array (SNP-A)-based karyotyping using Affymetrix Genome-Wide Human SNP Cytoscan HD. Signals at the top of each panel represent copy number status. The lower panel represents genotyping calls or the frequency of A and B alleles. Deletions are illustrated by a decrease in the copy number and a change in genotyping; interstitial deletion in chromosome 2, 7 and 20 are highlighted with a red box. The del(7)(q36) was not identified by metaphase cytogenetics. Normal copy number and three genotypes were observed in chromosome 19 by SNP-A. Chromosomes 9 and 22 presented normal copy number and genotypes by SNP-A. Bone marrow morphological, cytogenetics and molecular analysis of Case 1. (A) Hypercellular bone marrow fragment. The bone marrow was stained with Wright-Giemsa stain and the images show: (B) myeloblasts, (C) dyserythropoiesis with karyorrhexis, (D) dyserythropoiesis with binucleate erythroid precursor and nuclei/cytoplasm asynchronous maturation, (E) myeloid precursor with hypogranulation cytoplasm and (F) dysplastic small megakaryocytes with monolobed nuclei. The G-banded karyotype revealed 46,XY,t(9;22)(q34;q11),del(20)(q11)[2]/46,idem,inv(7)(q22q36)[6]/47,idem,inv(7)(q22q36),+19[12]; illustrative metaphase (G–I). The t(9;22)(q34;q11) is indicated with blue arrows, the del(20)(q11) is indicated with a green arrow, the inv(7)(q22q36) is indicated with a red arrow and the +19 is indicated with a black arrow. (J) Polymerase chain reaction for b2a2/b3a2 and e1a2 BCR-ABL1 transcripts – 1: ladder 50 base pairs (Life Technologies); 2: Case 1; 3: positive control for b2a2/b3a2 BCR-ABL1 transcript (p210); 4: positive control for e1a2 BCR-ABL1 transcript (p190); 5: negative control for b2a2/b3a2 and e1a2 BCR-ABL1 transcripts. (K) Single nucleotide polymorphism array (SNP-A)-based karyotyping using Affymetrix Genome-Wide Human SNP Cytoscan HD. Signals at the top of each panel represent copy number status. The lower panel represents genotyping calls or the frequency of A and B alleles. Deletions are illustrated by a decrease in the copy number and a change in genotyping; interstitial deletion in chromosome 2, 7 and 20 are highlighted with a red box. The del(7)(q36) was not identified by metaphase cytogenetics. Normal copy number and three genotypes were observed in chromosome 19 by SNP-A. Chromosomes 9 and 22 presented normal copy number and genotypes by SNP-A.](https://static.elsevier.es/multimedia/15168484/0000003900000004/v2_201911281025/S1516848417300993/v2_201911281025/en/main.assets/thumbnail/gr1.jpeg?xkr=ue/ImdikoIMrsJoerZ+w93OM6WmS6o9DeZl+SVh74uo=)