The bone marrow (BM) microenvironment plays a significant role in acute myeloid leukemia (AML) genesis and there is evidence that BM mesenchymal stromal cells (BMMSCs) can support leukemia progenitor cell proliferation and survival and provide resistance to cytotoxic therapies.
Hypothesis and methodNevertheless, currently unknown are the relevance of the spatial localization of AML cells relative to the BMMSCs and whether BMMSCs from patients with AML and healthy subjects have similar properties. To address these issues, we performed a differential gene expression analysis using RNA-sequencing data generated from healthy donors (HDs) and leukemic BMMSCs.
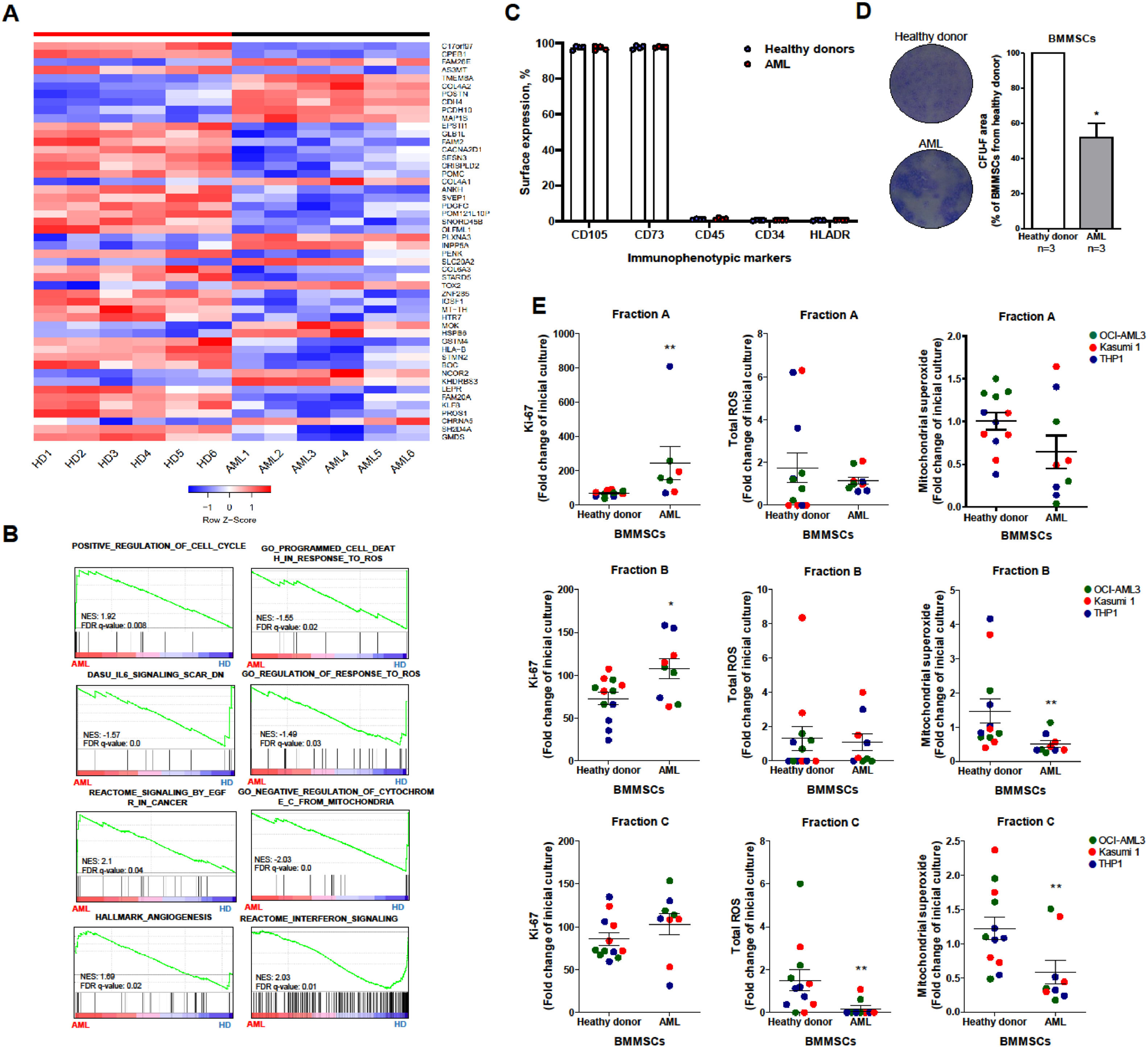
ResultsThe Gene Set Enrichment Analysis (GSEA) revealed that leukemic BMMSCs were associated with the terms “positive regulation of cell cycle”, “angiogenesis” and “signaling by the estimated glomerular filtration rate (eGFR)”, whereas healthy donor (HD)-derived BMMSCs were associated with “programmed cell death in response to the reactive oxygen species (ROS)”, “negative regulation of the cytochrome C from the mitochondria” and “interferon signaling”. Next, we evaluated the mitochondrial superoxide production in AML cells in a co-culture layered model. The superoxide production was reduced in leukemic cells in close contact (adhered to the surface or beneath the cell layer) with BMMSCs, indicating lower oxidative stress.
ConclusionTaken together, our results suggest that AML-derived BMMSCs are transcriptionally rewired and can reduce the metabolic stress of leukemic cells.
The bone marrow (BM) microenvironment is a heterotypic compartment composed of cellular (e.g., BM mesenchymal stromal cells (BMMSCs), endothelial cells, adipocytes, macrophages and fibroblasts) and extracellular (e.g., glycosaminoglycans, glycoproteins and collagen fibers) components that play a central role for self-renewal and differentiation of the hematopoietic stem cells (HSCs).1–4 The spatial distribution of cells within the BM microenvironment is believed to regulate the cell fate and ex vivo co-culture systems using BMMSCs as a feeder cell layer, suggesting that the BMMSC cell surface is the predominant site of the proliferation of HSCs, whereas the compartment beneath the BMMSC layer mimics the HSC niche Several studies have addressed the role of the BM microenvironment in leukemogenesis, especially regarding the contribution of BMMSCs.5,6 Nevertheless, presently unknown are the relevance of the localization of acute myeloid leukemia (AML) cells relative to the BMMSC layer and whether BMMSCs from patients with AML have effects similar to those from healthy subjects. To explore in more detail the biological process regulated on BMMSC cells from AML patients, we performed a differential gene expression analysis using a publicly available dataset of healthy and leukemic BMMSCs.5 Based on the functional genomics analysis, we evaluated the contribution of healthy donor (HD)- and AML-derived BMMSCs for the cell proliferation and reactive oxygen species (ROS) regulation in AML cell lines.
Material and methodsMicroarray and RNA sequencing (RNA-seq) data analysisThe RNA-seq data were obtained from the GSE92778 dataset, which was deposited in the GEO database (https://www.ncbi.nlm.nih.gov/geo).7 Genes that presented normalized counts equal to 0 for any replicates were excluded from the analysis.
Gene set enrichment analysis (GSEA) for AML-derived mesenchymal stromal cells (MSCs)Gene set enrichment analysis (GSEA) was performed using the Broad Institute software (http://software.broadinstitute.org/gsea/index.jsp). All genes from the RNA-seq of the GSE927787 were pre-ranked according to their differential expression (fold change) using the comparison of AML versus healthy donor-isolated MSCs. Enrichment scores (ESs) were obtained with the Kolmogorov-Smirnov statistic, tested for significance using 1000 permutations and normalized enrichment scores (NESs) to consider the size of each gene set. As suggested by the GSEA, a false discovery rate (FDR) cut-off of 25 % (FDR q-value < 0.25) was used.8 The limma-voom tool (http://usegalaxy.org) was used to examine differentially expressed genes and genes with ≥ 1log difference and adjusted p-value of < 0.05 were considered significant.9
Patient samplesFor functional assays, the BM aspirates were collected from de novo AML patients (n = 7; age range: 23 - 63 years; 1 favorable risk, 2 intermediate risk and 4 adverse risk according to the ELN2017 stratification)10 attended at the Ribeirão Preto Medical School Hospital das Clínicas. The diagnosis was performed following the criteria established by the World Health Organization.10,11 Healthy donors with no history of hematologic diseases were included (n = 9, age range: 26 - 54 years). The study was approved by the local Institutional Review Board (#7147/2005).
Isolation and characterization of BM-derived MSCsThe isolation of BMMSCs was performed by culturing BM mononuclear cells in the alpha minimum essential medium (α-MEM) containing 1 % penicillin/streptomycin and 20 % fetal bovine serum at 37 °C, with 5 % CO2. The obtained BMMSCs were characterized by immunophenotypic analysis (CD105, CD73, CD45, CD34 and HLA DR) and fibroblastic colony-forming unit (CFU F) capacity, according to the criteria recommended by the International Society for Cellular Therapy.12,13
Cell lines and experimental setupAll cell cultures were maintained in a humidified atmosphere at 37 °C with 5 % CO2. The OCI-AML3 (DSMZ - ACC582), Kasumi-1 (ATCC – CRL-2724) and THP-1 (ATCC – TIB-202) were cultured in the α-MEM with 20 % fetal bovine serum. The BMMSCs were seeded at the fourth passage and synchronized with 2 mM thymidine (Calbiochem, Darmstadt, Germany) for 16 h. Subsequently, AML cell lines were added in a 1:1 ratio and maintained in co-culture for 3 (defined as co-culture adaptation period) and 5 (defined as a co-culture final period) days. The separation of the leukemia cellular fractions was performed according to Jing et al.14, resulting in three different populations: non-adherent cells (fraction A), phase-bright cells (fraction B) and phase-dim cells (fraction C). In brief, the non-adherent cells were obtained by the aspiration of the co-culture (fraction A) supernatant. Following that, the MSC layer was gently washed twice with phosphate-buffered saline (PBS) to remove the superficially adherent cells that were performing transient interactions with the MSCs (fraction B). Finally, adherent cells (fraction C) were obtained by trypsinization of both leukemia and BMMSCs.14
To investigate the contribution of the BMMSCs to the cell proliferation and oxidative stress regulation of leukemia cells, membrane antigens were stained using the CD45 PerCP (Clone: 2D1; BD Biosciences, California, USA). In sequence, a fraction of the cells was permeabilized using the BD Perm III buffer (BD Biosciences) for 15 min and stained for the intracellular marker BV421 Ki-67 (Clone: 11F6) (Biolegend, USA) for cell proliferation, while a second fraction of the cells were stained with H2DCFDA for total ROS (5 µM) and MitoSOX® Red (ThermoFisher, USA) for mitochondrial superoxide production (50 nM). The acquisition and analysis were performed using a FACSCanto II flow cytometer and FlowJo analysis software (BD Biosciences), respectively (Supplemental Figure 1).
Statistical analysisAll p-values were two-sided with a significance level of 0.05. The Mann-Whitney or Kruskal-Wallis test was used to compare continuous variables. All statistical analyses were performed using the statistical package for the social sciences (SPSS) 19.0 and R 3.3.2 (The CRAN project, www.r-project.org) software.
ResultsInitially, in exploring the differential biological pathways enriched in AML-derived MSCs, the differential gene expression analysis indicated a homogeneous genetic distribution within the HD- and AML-derived BMMSC groups. The differentially expressed genes (adjusted p-value = 0.05) are illustrated in Figure 1A. Subsequently, GSEA associated AML-derived BMMSCs with the terms “positive regulation of cell cycle”, and “signaling by eGFR in cancer”, whereas HD-derived BMMSCs were associated with the terms “programmed cell death in response to ROS”, “Negative regulation of cytochrome C from mitochondria” and “Interferon signaling” (Figure 1B) These results suggested that AML-derived BMMSCs were transcriptionally rewired to provide better support for leukemic blasts.
Acute myeloid leukemia-derived bone marrow mesenchymal stromal cells improve cell proliferation and reactive oxygen species regulation in leukemia cells. (A) The heatmap was constructed using Morpheus and summarizes the expression of the differentially expressed genes (p < 0.05) across samples from healthy donor (HD)-derived bone marrow mesenchymal stromal cells (BMMSCs) (n = 4; HD1–4) versus acute myeloid leukemia (AML)-BMMSCs (n = 4; AML1–4). The color intensity represents the &#¿;-score within each row. (B) GSEA on a pre-ranked gene list based on the leading-edge genes for AML- and HD-derived BMMC expression. Genes were ranked based on Pearson's correlation with highly expressed genes in AML-derived BMMSCs. The normalized enrichment score (NES) and false discovery rate (FDR) were used for significance. (C) Bar graphs represent the mean ± SD of the percentage CD105, CD73, CD45, CD34 and HLA-DR cells of HD-derived (n = 4) and AML-derived BMMSC (n = 4) samples. (D) For fibroblastic colony forming unit (CFU-F) assay, BMMSCs were seeded in 6-well plates at a confluence of 500 cells/wells. After 15 days, CFU-F was detected by crystal violet staining. A minimum number of 50 cells grouped along the well were considered a colony. Colony images illustrate an experiment for HD-derived and AML-derived BMMSCs and the graphs represent the mean ± SD of three independent samples for each group. The p-values are indicated in the graphs; *p < 0.05; Student´s T-test. (E) Leukemia cells (gated CD45 cells) from fraction A (non-adherent population), fraction B (phase-bright population) and fraction C (adherent) were submitted to Ki-67, reactive oxygen species (ROS) and mitochondrial superoxide evaluation by flow cytometry, as indicated. Data were expressed as fold-change of the co-culture adaptation period (three days after mixing leukemia and BMMSCs for a cellular organization in fractions) and the final co-culture period (five days of incubation). The p-values and cell lines are indicated. *p < 0.05; **p < 0.01, Mann Whitney test.
Subsequently, we confirmed that BMMSCs were successfully cultivated under the conditions described above and no difference in the immunophenotype was detected between the BMMSCs derived from healthy donors those from and AML patients (Figure 1C). The fibroblastic colony formation unit evaluation (CFU-F) revealed that AML-derived BMMSCs presented a lower proliferative capacity, compared to HD-derived BMMSCs (Figure 1D). These data corroborate previous reports.15,16 Nevertheless, the effect on proliferation was dependent on the distribution of cells in the co-culture system. In the final culture (Day 5), leukemia cells from fractions A and B presented higher Ki-67 staining in the presence of AML-derived BMMSCs, compared to the HD-derived ones (p < 0.05), indicating higher proliferative potential (Figure 1E).
Regarding their supportive functions to leukemic blasts, the AML-derived BMMSCs were able to promote the survival of malignant cells throughout the ROS induction by reducing cellular damage, decreasing apoptosis, inducing quiescence, or a combination of these. The AML cells from fractions B and C presented reduced mitochondrial superoxide production in the presence of AML-derived BMMSCs, compared to the HD-derived ones, indicating lower oxidative stress (p < 0.05). In fraction C, Ki-67 levels evaluated by the mean intensity of fluorescence (MFI) were similar among leukemia cells, independently of the origin of the BMMSCs, but a reduction in the total ROS was observed in leukemia cells in the presence of AML-derived BMMSCs (p < 0.05; Figure 1E). Taken together, these results suggested that the close contact between the AML blasts and BMMSCs can reduce the metabolic stress of leukemic cells, protecting them from environmental stress.
DiscussionHerein, we functionally demonstrated the impact of BMMSCs isolated from healthy donors and those from AML patients on cellular processes identified in the functional genomic analysis, cell proliferation and ROS regulation, which was evaluated using in vitro AML cellular models. It has been reported that BMMSCs from AML present less proliferation, clonogenic capacity and support for normal HSCs.16,17,18 These findings reinforce the theory of the AML cells promoting modulation in the BM microenvironment during the disease development in a manner which causes it to acquire a superior capacity for supporting AML cells, compared to physiologic conditions. On the other hand, their role in maintaining leukemia cells is still an active field of study.19,20 Previous studies have reported that during leukemogenesis, leukemia blasts secrete various cytokines and growth factors, such as SCF, IL-4, IL-6 and IL-1b, promoting a change in the cell plasticity of the BM niche and assisting in the establishment of the disease.20 In agreement, we observed a lower proliferative capacity of the BMMSCs from AML, compared to those from healthy donors.
Ludin et al.21 reported that AML-derived BMMSCs have a superior ability to reduce the ROS in leukemic cells, probably mediated by a higher mitochondrial exchange capacity, compared to HD-BMMSCs. In the present study, the reduction of the total ROS observed in fraction C may indicate that AML-derived BMMSCs improve the quiescence of leukemia cells in close contact with BMMSCs, further protecting these cells from stressors, such as chemotherapy.5
Our study opens perspectives on the participation of BMMSCs in the proliferation of different subpopulations of leukemia cells; however, it has limitations. Due to methodological limitations, only the Ki-67 marker was used to assess the cell proliferation rate.22 Despite this molecular marker being widely accepted, used and validated as a proliferation marker in experimental models and in diagnostic medicine, future studies should investigate markers of cell cycle progression, differentiation, quiescence, apoptosis and senescence.
ConclusionIn summary, the present study confirms previous findings that BMMSCs isolated from AML patients have impaired proliferative capacity, improve leukemia cell proliferation and protect leukemia cells from metabolic stress.
This study was supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, Grant #2013/08135-2, #2021/11606-3) and the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). D.A.P-M. received a fellowship from FAPESP (Grant #2017/23117-1).