The aberrant expression of the inhibitor of DNA binding (ID1) gene has been frequently associated with the leukemogenesis and prognostication acute myeloid leukemia (AML), although its clinical importance has never been investigated in patients treated outside well-controlled clinical trials.
MethodsUsing quantitative real-time polymerase chain reaction, we investigated the role of the ID1 expression in the clinical outcomes of non-selected patients with acute myeloid leukemia treated in a real-life setting.
ResultsOverall, 128 patients were enrolled. Patients with high ID1 expression had a lower 3-year overall survival (OS) rate of 9%, with the 95% confidence interval (95%CI) at 3 to 20%, compared to patients with a low ID1 expression (22%, 95%CI: 11 - 34%) (p = 0.037), although these findings did not retain significance after adjustment (hazard ratio (HR): 1.5, 95%CI: 0.98 - 2.28; p = 0.057). The ID1 expression had no impact on post-induction outcomes (disease-free survival, p = 0.648; cumulative incidence of relapse, p = 0.584).
ConclusionsAlthough we are aware thar our data are confronted with many variables that cannot be fully controlled, including drug unavailability, risk-adapted treatment, comorbidities and the time from diagnosis to treatment initiation, we are firm believers that such an initiative can provide more realistic data on understudied populations, in particular those from low- and middle-income countries.
The aberrant expression of the inhibitor of the DNA binding (ID1) gene has been frequently associated with the initiation and progression of human tumors.1,2 Particularly in acute myeloid leukemia (AML), a growing body of evidence suggests that the ID1 overexpression can impact on both the AML leukemogenesis3,4 and prognostication,5–7 although these findings remain controversial. Most importantly, the clinical importance of the ID1 expression in patients treated outside well-controlled clinical trials has never been evaluated, which means that its potential prognostic value in a real-life context remains unknown. Hence, we investigated the role of the ID1 expression on clinical outcomes of non-selected AML patients treated at a single center in northeastern Brazil.
Material and methodsPatients and study designWe performed a retrospective cohort study investigating the clinical impact of the ID1 expression on patients with acute myeloid leukemia (AML) at diagnosis. Between January 2011 and November 2019, 128 consecutive patients were diagnosed with AML at three Brazilian reference centers specialized in AML treatment located in northeastern Brazil. The AML diagnosis was determined by cytomorphology, immunophenotyping and genetic studies, in accordance with the WHO criteria. In this study, only patients diagnosed with the de novo AML (128 patients) were included, while those with acute promyelocytic leukemia (31 patients), therapy-related AML (18 patients), or a previous history of myelodysplastic syndrome (14 patients) were excluded. An overview of patient characteristics according to the ID1 expression can be found in Table 1. All patients or their relatives gave their written informed consent for scientific evaluations. The study was approved by the Internal Review Board (#7147/2005) and adhered to the tenets of the Declaration of Helsinki.
Treatment protocolFor six of the 128 patients, there was no available information on the treatment procedures or their respective follow-ups. Consequently, they were excluded from induction and post-induction analyses. The majority of the patients (91 patients, 74%) were intensively treated with cytotoxic chemotherapy. The induction therapy consisted of daunorubicin (60 or 90 mg/m 2 daily for 3 days) and cytarabine (100 or 200 mg/m 2 daily for 7 days) or thioguanine, cytosine arabinoside and daunorubicin,8 followed by two or three cycles of consolidation therapy with high-dose cytarabine (1.5 g/m 2 or 3 g/m 2 for 3 days). The post-remission therapy based on autologous or allogeneic transplantation was not performed in only ten patients. The complete remission (CR) was assessed by bone marrow examination 28 days after each course of chemotherapy. Patients who did not achieve complete remission after one course of chemotherapy, received a second course. Of the 31 patients who did not receive intensive treatment, 24 were 60 years old or over. These patients were treated with low-dose ARA-C, in combination with etoposide, thioguanine and idarubicin, or exposed to palliative care.
Cytogenetic and molecular analysesAll materials used for genetic analyses were obtained at diagnosis and were processed in the reference laboratories of each participating center. The cytogenetic analysis was performed from bone marrow aspirates according to the standard techniques for chromosomal banding.
The real-time quantitative polymerase chain reaction (RT-qPCR) methodology was used to determine the ID1 transcript levels. Following total RNA extraction, RT-qPCR assays were performed in duplicate, using sample-derived complementary DNA (cDNA) on MicroAmp optical 96-well plates, using the QuantStudio™ 5 Dx Real-Time PCR System (Applied Biosystems, Foster City, CA), with the human GAPDH pre-developed Taqman assay as the endogenous control. The ID1 gene expression was determined using the TaqMan Gene Expression Assay (Hs03676575_s1; Applied Biosystems) following the manufacturer's instructions. The gene expression of the ID1 was calculated relative to a reference cDNA obtained from the Ontario Cancer Institute-Acute Myeloid Leukemia-3 (OCI-AML3) cell line. Importantly, the same reference cDNA was used as an internal control in all experiments to ensure that the results of different experiments could be comparable. The ID1 relative gene expression was quantified using the Δ quantification cycle (Cq) method and the results were expressed using the 2−ΔΔCt.
Statistics analysis and clinical endpointsDescriptive analyses were performed for patient baseline features. To define the optimal cutoff for ID1 transcript levels, the total cohort was initially divided into quartiles (Q) according to the ID1 expression (as fold change). Based on the survival curves, quartiles with similar event probabilities would be grouped. Unfortunately, there was no clear distinction between groups (Supplemental figure 1). Subsequently, we used the receiver operating characteristic (ROC) curve analysis to define the optimal cut-off point for the ID1 expression, but because of a limited number of patients, the performance of the ROC curve analysis was unsatisfactory (area under the curve: 0.525, 95% confidence interval (95%CI): 0.411 - 0.639). Based on these analyses, we opted to adopt the median value of the ID1 expression (as fold change) as the cut-off to dichotomize patients into low and high ID1 transcript levels. The Fisher's exact test or Chi-square test, as appropriate, was used to compare categorical variables. The Kruskal-Wallis test was used to compare continuous variables. Details of the statistical analysis and clinical endpoints were described elsewhere.9 All p-values were two-sided, with a significance level of 0.05. All calculations were performed using the Stata Statistic/Data Analysis version 12 (Stata Corporation, USA) and the R 3.3.2 (The CRAN project, www.r-project.org) software.
Public datasetsThe patient clinical features, mutational and transcriptome data from the Cancer Genome Atlas Program (TCGA)10 were obtained from the site http://cancergenome.nih.gov. For the RNAseq analysis, the normalization of the counts by reads per kilobase million (RPKM) was used for the expression analysis and for the microarray data, the normalized fluorescence of the probe 208937_s_at.
The gene set enrichment analysis (GSEA) was conducted using the preranked GSEA11 from the TCGA cohort data. The gene set database (all gene sets from the hallmark collection and gene sets from the curated collection with the homo sapiens organism) used for the analysis was obtained from the molecular signatures database (MsigDB) v6.2. The input data for the pre-ranked GSEA was the ranked list of genes based on the Pearson's correlation coefficient for all genes after filtering (19,882 genes).
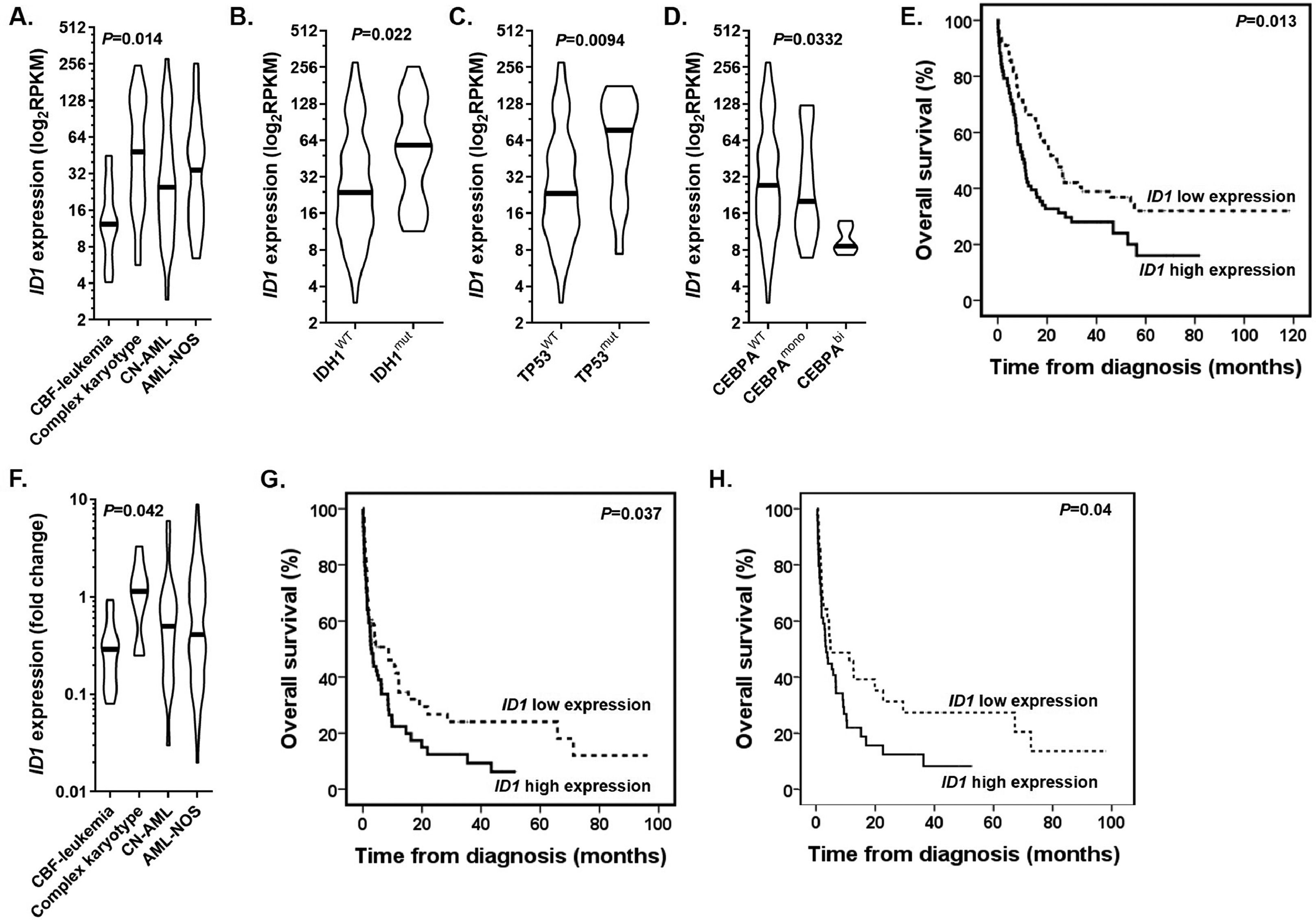
ResultsTo test the feasibility of the study, we took advantage of The Cancer Genome Atlas database (154 non-acute promyelocytic leukemia patients, hereafter called “training cohort”) and explored the association of the ID1 expression with clinical and laboratory features. After classifying patients according to cytogenetic abnormalities at diagnosis, we noticed that the ID1 expression was significantly lower in patients with the core binding factor (CBF) leukemia (p = 0.014; Figure 1A). Moreover, the ID1 was overexpressed in the IDH1- (p = 0.022; Figure 1B) and TP53-mutated patients (p = 0.0094; Figure 1C), while biallelic CEBPA-mutated patients presented lower ID1 expression (p = 0.0332; Figure 1D). In sequence, we determined the prognostic impact of the ID1 expression on the patient treatment outcomes. Using the median value of the ID1 expression, patients were dichotomized into low and high expression groups. In the univariate analysis, a high ID1 expression was associated with the poor 5-year overall survival (OS) rate (p = 0.013; Figure 1E). However, the ID1 expression did not retain its prognostic impact after the adjustment for age (continuous variable), leukocyte counts (continuous variable) and cytogenetic risk stratification (Supplemental Table 1). Furthermore, there was no impact of the ID1 expression on post-remission outcomes (disease-free survival, p = 0.615; cumulative incidence of relapse, p = 0.52). The Supplemental Figure 2 shows the Gene Set Enrichment Analysis pre-ranked comparing the transcriptome of patients with high and low expression of the ID1 gene. The GSEA revealed that samples with high ID1 expression were significantly enriched for gene sets associated with FLT3-ITD mutated AMLs, cell proliferation, ribosomal biogenesis and translation. Patients with low ID1 expression were enriched for gene sets characteristic for AMLs with the mixed lineage leukemia (MLL) translocations and CBF-leukemia rearrangements, in particular the CBFB-MYH11 fusion gene.
(A) The ID1 expression according to the cytogenetics abnormalities at diagnosis of AML patients from the training cohort. (B-D) The ID1 expression according to the mutational status of (B) the IDH1, (C) TP53 and (D) CEBPA genes. (E) The comparison of OS curves for patients from the training cohort with low and high ID1 expression. (F) The ID1 expression according to the cytogenetics abnormalities at diagnosis of AML patients from the real-life cohort. (G) The comparison of OS curves for patients from the real-life cohort with low and high ID1 expression (entire cohort) (H) and patients who were intensively treated with chemotherapy (91 patients).
To translate these findings into a resource-constrained scenario, we enrolled 128 adult de novo AML patients diagnosed and treated in northeastern Brazil (here referred to as “resource-constrained cohort”). Details for treatment protocols and the inclusion criteria were published elsewhere.12 All samples used were obtained at diagnosis from bone marrow aspirates. Ethical approval was obtained from the local Research Ethics Board (CAAE: 77527717.4.0000.5208). In accordance with the Declaration of Helsinki, informed written consent for the sample collection, as well as permission for its use in the research, was obtained from patients or their relatives.
Figure 1F shows that patients with CBF-leukemia from the resource-constrained cohort also exhibited a low ID1 expression, compared to other cytogenetic risk groups (p = 0.042). Here, we opted to adopt the same strategy of dichotomization used in the training cohort. Based on the median value of the ID1 expression (median value: 0.485, range: 0.1 - 8.9), high ID1 patients had low platelet counts (p = 0.04) and higher levels of lactate dehydrogenase (p = 0.021) (Table 1). Of the 128 enrolled patients, 6 (5%) patients did not have available information on the treatment procedures or their respective follow-ups and were considered ineligible for the induction therapy analyses. The median follow-up was 35 months (95%CI: 30 –40 months) (Shouldn't these be percentages??? ) and the estimated 3-year OS rate was 15% (95%CI: 8 - 23%). Overall, 58/122 (47%) patients achieved complete remission. The ID1 expression had no impact on the CR achievement (p = 0.276). Patients with a high ID1 expression had a lower 3-year OS (9%, 95%CI: 3 - 20%) compared to patients with a low ID1 expression (22%, 95%CI: 11 - 34%) (p = 0.037) (Figure 1G). The best-fitted multivariate Cox proportional hazards model for the OS was the age (continuous variable), leukocyte counts (continuous variable), and ID1 expression (Supplemental Table 2). This model showed that the ID1 expression was not independently associated with a poor OS (hazard ratio (HR): 1.5, 95%CI: 0.98 - 2.28; p = 0.057). The ID1 expression had no impact on the disease-free survival (p = 0.648) or cumulative incidence of relapse (p = 0.584).
Finally, we opted to evaluate the impact of the ID1 expression separately in patients submitted to intensive chemotherapy (91 patients). Although the CR achievement rate was higher in patients with a low ID1 expression (58% versus 44%), this difference was not significant (p = 0.17). On the other hand, patients with a high ID1 expression had a lower 3-year OS (4%, 95%CI: 4 - 27%), compared to patients with a low ID1 expression (33%, 95%CI: 18 - 50%) (p = 0.04) (Figure 1H). The ID1 expression had no impact on the disease-free survival (p = 0.604) or cumulative incidence of relapse (p = 0.74).
DiscussionThree issues require particular attention. Contrary to Tang et al., 5 we and others6,7 have failed to demonstrate an independent association between high ID1 expression and poor induction outcomes in AML. The possible reasons for this disparity are multifactorial and may range from patient-related features (such as age and ethnic differences) to differences in treatment protocols, in particular post-remission therapy. Furthermore, we cannot rule out that methodological differences between studies (different standards for the ID1 quantification or different definitions for “high” and “low” ID1 expression) could lead to different results and conclusions. These arguments are frequently used to justify differences between studies. But because our study has included consecutive and non-selected patients treated outside well-controlled clinical trials, an additional topic needs to be addressed. Our data are confronted with many variables that cannot be fully controlled, including drug unavailability, risk-adapted treatment, comorbidities, and time from diagnosis to treatment initiation.12 Despite this obvious limitation involving clinical studies conducted in a real-world setting, we are firm believers that such an initiative can provide more realistic data on understudied populations, in particular those from low- and middle-income countries.
Regardless of whether the ID1 is an independent prognostic factor in AML or not, an outstanding question is whether its implementation could improve the current scheme for the AML risk stratification.13 Based on its biological importance, it is conceivable that the ID1 gene or its encoded protein may be useful in future revised versions of the scheme for the AML risk stratification.14 Initially, the encoded Id1 protein seems to be a key transcriptional regulator of the hematopoietic stem cell (HSC) lineage commitment and the absence of the Id1 may compromise the self-renewal capacity of HSCs.15 In human tumors, the Id1 protein can control cell proliferation, self-renewal capacity of cancer stem cells and disease aggressiveness via myelocytomatosis (Myc).16 Furthermore, the ID1 overexpression induces cell proliferation and invasion and also protects cells against drug-induced apoptosis.17 In a hematological context, the ID1 overexpression is able to immortalize myeloid progenitors in vitro and promote myeloproliferative-like phenotype in vivo.18 Conversely, its deletion in the hematopoietic stem cell decreases cell proliferation, mitochondrial biogenesis, metabolic activity and ribosomal biogenesis.19 It is important to note that the encoded Id1 protein is a common downstream target of constitutively activated oncogenic tyrosine kinase in AML4 and can be upregulated during the CEBPA-induced myeloid differentiation.20 Additionally, the forced expression of the MLL-AF9 fusion gene in fetal liver cells leads to an increase in the ID1 expression and the rapid development of AML with maturation in a fetal liver transplant model.3 Together, these pieces of evidence support two main conclusions. First, the encoded Id1 protein plays an important role in the pathophysiology of human tumors and could be useful in understanding the pathophysiology of AML. Second, the deregulated ID1 expression does not seem to be a primary genetic event in leukemic blasts, but a surrogate marker depending on other genetic aberrations. If so, it is conceivable that the ID1 overexpression may be associated with specific AML subtypes, which could explain the downregulation of the ID1 expression in CBF-leukemia reported here in both training and resource-constrained cohorts.
ConclusionIn summary, the ID1 expression was associated with the induction outcomes in patients treated outside well-controlled clinical trials, although not in an independent manner. With respect to its potential use in clinical practice (either as a potential therapeutic target or prognostic factor), further studies are necessary to validate this hypothesis.
Author contributionsA.S.L. and M.F.B. performed experiments, collected, analyzed and interpreted data and drafted the manuscript. A.S.L., R.M.F. and A.R.L-A. performed the statistical analyses, interpreted the data and drafted the manuscript. A.M-A, I.W., B.L.S., M.L.S-B, P.L.F-N, M.M.L. F.S-A. J.L.C-S, D.A.P-M., M.A.B obtained patient samples, updated the clinical data, collected, analyzed and interpreted data and drafted the manuscript. I.W. and A.R.L-A. conceived and designed the study and reviewed the manuscript. A.R.L-A. gave the final approval of the version to be submitted.
We gratefully acknowledge financial support from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) grant # 406570/2018-3. A.M-A received a fellowship from the CNPq (grant #160370/2021-3). M.LS-B. received a fellowship from the Fundação de Amparo a Ciência e Tecnologia de Pernambuco (Grant # IBPG-1108-2.02/21).