Myelodysplastic syndrome (MDS) constitutes a heterogeneous group of hematological malignancies characterized by ineffective hematopoiesis, presence of cytopenias and high risk of progression to acute myeloid leukemia (AML).1–3 Chromosomal abnormalities can be identified by conventional cytogenetics (G-banding) in up to 50% of MDS patients; this is considered the most important marker of prognosis.3,4
Clonal evolution is defined as an abnormal clone in a patient whose karyotype was normal or an additional aberration in a patient whose karyotype was abnormal at first examination.5,6 Different combinations of chromosomal alterations and somatic point mutations contribute to the large clinical–pathological spectrum of MDS.7 Whole-arm translocations (WATs) are described as a single and rare cytogenetic abnormality in hematologic malignancies, suggesting that these may be primary changes.7,8 The main consequence of WATs is the genomic instability resulting from the gain or loss of whole chromosome arms.8
The der(1;7)(q10;p10) is an unbalanced translocation associated with myeloid disorders.9,10 This translocation is a product of mitotic recombination within centromeric regions, which lead to allelic imbalances of a trisomy 1q and a deletion 7q.10 This chromosomal abnormality is relatively rare in MDS; it has been reported in 1–3% of MDS patients and in 1–2% in AML patients.8–10 The presence of rare cytogenetic abnormalities occurs most frequently in patients with complex karyotypes; furthermore, the prognostic classification of rare abnormalities remains a challenge.10,11 The aim of this report is to show a rare case of an MDS patient who presented der(1;7)(q10;p10) during clonal evolution and discuss the importance of sequential karyotype analysis in the follow up of MDS.
Case ReportA 65-year-old man looked for medical assistance due to severe thrombocytopenia. At physical examination, the patient did not present bruises and denied bleeding. The patient reported a history of smoking and occupational exposure to pesticides for decades. A complete blood count (CBC) revealed hemoglobin 12.4g/dL, leukocytes 2.098×109/L, neutrophils 0.482×109/L and platelets 0.064×109/L. The bone marrow aspirate showed a discrete dysplasia (<10%) in the three cell lines. Bone marrow biopsy demonstrated a normocellular marrow with dysplasia of the erythroid and granulocytic lines. The first cytogenetic analysis showed: 46,XY[5]. After six months of follow-up another cytogenetic analysis showed the absence of metaphases. The diagnosis of MDS with multilineage dysplasia (MDS-MLD) was established according to the World Health Organization (WHO) 2016 classification.12
By the fourth year of follow up, another cytogenetic examination was performed, with the karyotype: 46,XY[20]. During the fifth year of follow-up the CBC showed hemoglobin 9.2g/dL, leukocytes 0.692×109/L, neutrophils 0.166×109/L and platelets 0.081×109/L. At this time, a new cytogenetic analysis was performed: 46,XY,der(7)t(1;7)(q10;p10)[18]/46,XY[2] (Figure 1). Bone marrow aspirate showed dysplasias in the erythroid, granulocytic and megakaryocytic lines with the presence of 6% of blasts. The diagnosis of MDS with excess blasts-1 (MDS-EB-1) was established according to the WHO classification 12 and the Revised International Prognostic Scoring System (IPSS-R)13 was high.
DiscussionThe detection of chromosomal abnormalities by G-banding is one of the strongest prognostic parameters in MDS and has a fundamental role in its classification.14 Patients with complex karyotypes and clonal evolution have a higher risk of AML transformation.15,16 The most common chromosomal abnormalities in MDS are del(5q), del(7q)/monosomy 7 and trisomy 8, nevertheless, non-informative karyotypes are still found in up to 20% of the cases.14–16 Der(1;7)(q10;p10) is a nonrandom translocation in MDS, formed due to similarity between the centromeric region sequences of chromosomes 1 and 7.17 In a study of 131 patients with hematological malignancies, the loss of 7q and gain in 1q regions were identified in more than 10% of the cases.18 Several studies reported the association of therapeutic or environment exposure to toxic substances with der(1;7)(q10;p10) in about half of the cases.16–18
Hsiao et al. evaluated 19 cases of MDS and four cases of AML with der(1;7)(q10;p10).19 The cytogenetic results showed that ten patients (43.5%) had der(1;7) as the single chromosomal alteration. In addition, eight patients (34.8%) reported a history of exposure to genotoxic agents.19 It is interesting to note that our patient has a history of occupational exposure to pesticides and reported being a smoker for decades; both these exposure factors may have contributed to the clonal evolution reported herein.19
Zhang et al. performed target next-generation sequencing (NGS) on 22 MDS cases with der(1;7)(q10;p10) and 32 MDS patients with -7/del(7q) to better understand the somatic mutation spectrum of these entities.20 The most frequently mutated genes were RUNX1 (40.9%) in der(1;7) and TP53 (28.1%) in -7/del(7q) patients.20 In addition, TP53 mutations were only detected in -7/del(7q) patients and not found in patients with der(1;7).20 These data are interesting because some authors speculated that der(1;7)(q10;p10) represents a ‘karyotypic variant’ of 7(-7/7q-).17–20 However, Zhang et al. also showed that these groups have a different mutational profile and that der(1;7)(q10;p10) is associated with a high frequency of mutations in RUNX1.20 Detailed characterization of cytogenetic findings and the genes affected by these abnormalities will further improve our knowledge of the cellular events that lead to MDS and enhance our understanding of complex biology and the dynamic nature of this disease.21
Our patient performed four cytogenetic analyses since his follow-up was started with the first examination being normal even though only five metaphases were analyzed, the next reported absence of metaphases, then normal again and the last one showed the presence of der(1;7). The fluorescent hybridization in situ (FISH) technique could be performed in cases with a low mitotic index and an absence of metaphases in order to add important information to the diagnosis and evaluation of the patient.22 Thus, FISH has a real benefit in from 1 to 32% depending on the risk of the MDS patients.22
Sequential karyotype analysis is usually recommended when the patient shows any new cytopenia or worsening of the cytopenia.22,23 We also suggest that karyotyping should be repeated annually. We prospectively evaluated 50 MDS patients at diagnosis, and after six and 12 months of follow-up.23 As a result, chromosomal abnormalities were detected in 19.5% (9/46) of the patients at diagnosis, 27% (10/37) at the end of the sixth month and 35.4% (11/31) at the end of the twelfth month of follow-up.23
ConclusionOur results show the importance of sequential karyotype analysis in the follow-up of MDS in order to observe possible clonal evolution which is a frequent event in MDS and, similar to the development of point mutations, reflects the heterogeneous and dynamic nature of this disease.23
Conflicts of InterestThe authors declare no conflicts of interest.
This study was conducted with partial support from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq)


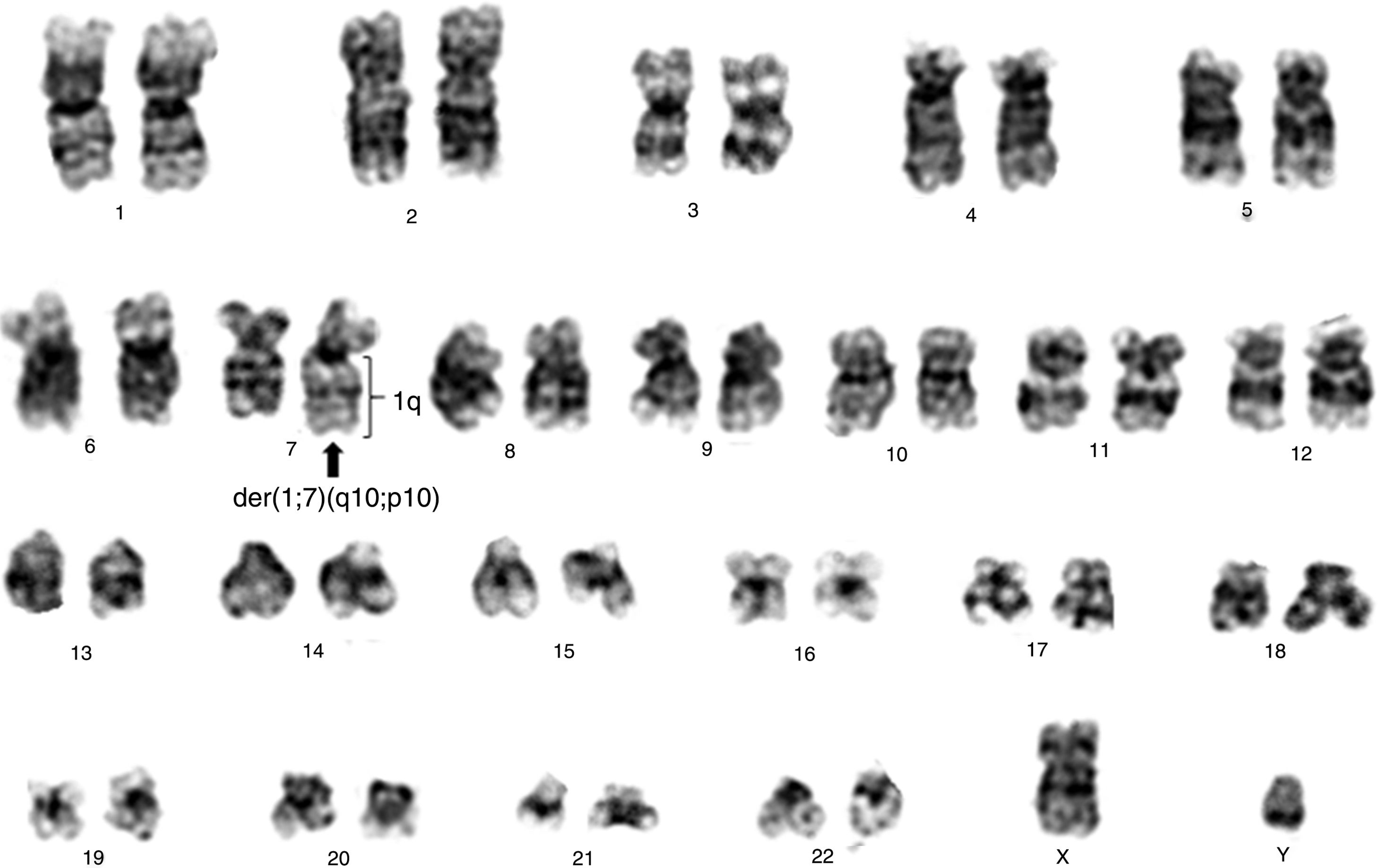
![Conventional cytogenetics study of patient showing 46,XY,der(7)t(1;7)(q10;p10)[18]/46,XY[2]. Conventional cytogenetics study of patient showing 46,XY,der(7)t(1;7)(q10;p10)[18]/46,XY[2].](https://static.elsevier.es/multimedia/25311379/0000004100000001/v2_201911290921/S2531137918301020/v2_201911290921/en/main.assets/thumbnail/gr1.jpeg?xkr=ue/ImdikoIMrsJoerZ+w93OM6WmS6o9DeZl+SVh74uo=)





