A 67-year-old male presented with a four-week history of weakness. The complete blood count showed: hemoglobin: 5.1g/dL, leukocytes 182.55×103/μL (23% blasts and 34% monocytes) and platelets: 31×109/L. A bone marrow aspirate showed 50.4% of myeloid myeloperoxidase (MPO)-blast cells and 42.4% of dysplastic granulocytic-monocytic series, with alpha-naphthyl acetate esterase positivity in 30% of total nucleated cells. Flow cytometry identified two distinct aberrant blasts (CD4-CD7-CD11c-CD13-CD34-CD117-HLA-DR-cMPO+) and myeloid/monocytic (CD14-CD33-CD35-HLA-DR-CD11b+) populations. Karyotyping showed monosomy 7 and additional material in the long arm of chromosome 2 (Figure 1A). Acute myeloid leukemia (AML)-M4 (FAB classification) or AML with myelodysplasia-related changes (WHO 2008 classification) was diagnosed. Patient died two months after without response to therapy.
Apart from karyotyping, other molecular genetic techniques can detect gains and losses of genomic material.1–3 In this case, the additional material in chromosome 2 was elucidated and chromosome 7 monosomy was confirmed using fluorescence in situ hybridization, multiplex ligation-dependent probe amplification and single nucleotide polymorphism-array methodologies (Figures 1B, 2A and B).
(A) MLPA with SALSA MLPA probemix P144-A2 (above) and P145-A2 (below) kits (MRC-Holland, Amsterdam, The Netherlands). Dosage quotient of the patients’ probes in relation to a control group. Probes positioned below the 0.65 limit indicate deletion; probes positioned above the 1.35 limit indicate duplication. Probes in red indicate monosomy 7 (deletion of all probes of chromosome 7) and probes in black are normal for the other chromosomes studied in these kits. (B) SNP-A (CytoScanHD Array; Affymetrix, Santa Clara, USA): Diagrams generated by ChAS (Affymetrix) for chromosome 2 (above) and chromosome 7 (below). In “Copy number state”, line in 2.00 indicates a normal copy number (diploid), line in 1.00 indicates a deletion and line in 3.00 indicates a duplication/gain. Lines with intermediate values between 1.00 and 2.00; 2.00 and 3.00 indicate mosaicism. In “Allele Difference”, three lines indicate a normal genotype (AA, AB and BB alleles), two lines indicate deletion (A and B alleles) or loss of heterozygosity (AA and BB alleles) and four lines indicate duplication (AAA, AAB, ABB and BBB alleles). SNP-A revealed the result arr[hg19] 2p25.3p16.2(12770_54276429)x3[0.8],2q35q37.3 (219576639_242783384)x1,(7)x1 where there was a partial duplication of the short arm of chromosome 2 (A and C), presenting clonal mosaic data of the duplicated region in 80% of the cells, and partial deletion of the long arm of chromosome 2 (B and D). This result can explain add (2) (q35) present in the karyotype, indicating the origin of the additional material. In addition, the monosomy of chromosome 7 was confirmed.
The authors declare no conflicts of interest.


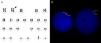

![(A) Karyotype (G-band): 45,XY,add(2)(q35),-7[20]. (B) FISH (Del (7q) Deletion Probe, ref: RU-LPH 025; Cytocell, Cambridge, UK): Deletion of RELN gene (chromosome 7) in 96% of the analyzed nuclei. The absence of a green and a red signal may indicate the monosomy of the chromosome. (A) Karyotype (G-band): 45,XY,add(2)(q35),-7[20]. (B) FISH (Del (7q) Deletion Probe, ref: RU-LPH 025; Cytocell, Cambridge, UK): Deletion of RELN gene (chromosome 7) in 96% of the analyzed nuclei. The absence of a green and a red signal may indicate the monosomy of the chromosome.](https://static.elsevier.es/multimedia/15168484/0000003900000004/v2_201911281025/S1516848417300920/v2_201911281025/en/main.assets/thumbnail/gr1.jpeg?xkr=ue/ImdikoIMrsJoerZ+w93OM6WmS6o9DeZl+SVh74uo=)
![(A) MLPA with SALSA MLPA probemix P144-A2 (above) and P145-A2 (below) kits (MRC-Holland, Amsterdam, The Netherlands). Dosage quotient of the patients’ probes in relation to a control group. Probes positioned below the 0.65 limit indicate deletion; probes positioned above the 1.35 limit indicate duplication. Probes in red indicate monosomy 7 (deletion of all probes of chromosome 7) and probes in black are normal for the other chromosomes studied in these kits. (B) SNP-A (CytoScanHD Array; Affymetrix, Santa Clara, USA): Diagrams generated by ChAS (Affymetrix) for chromosome 2 (above) and chromosome 7 (below). In “Copy number state”, line in 2.00 indicates a normal copy number (diploid), line in 1.00 indicates a deletion and line in 3.00 indicates a duplication/gain. Lines with intermediate values between 1.00 and 2.00; 2.00 and 3.00 indicate mosaicism. In “Allele Difference”, three lines indicate a normal genotype (AA, AB and BB alleles), two lines indicate deletion (A and B alleles) or loss of heterozygosity (AA and BB alleles) and four lines indicate duplication (AAA, AAB, ABB and BBB alleles). SNP-A revealed the result arr[hg19] 2p25.3p16.2(12770_54276429)x3[0.8],2q35q37.3 (219576639_242783384)x1,(7)x1 where there was a partial duplication of the short arm of chromosome 2 (A and C), presenting clonal mosaic data of the duplicated region in 80% of the cells, and partial deletion of the long arm of chromosome 2 (B and D). This result can explain add (2) (q35) present in the karyotype, indicating the origin of the additional material. In addition, the monosomy of chromosome 7 was confirmed. (A) MLPA with SALSA MLPA probemix P144-A2 (above) and P145-A2 (below) kits (MRC-Holland, Amsterdam, The Netherlands). Dosage quotient of the patients’ probes in relation to a control group. Probes positioned below the 0.65 limit indicate deletion; probes positioned above the 1.35 limit indicate duplication. Probes in red indicate monosomy 7 (deletion of all probes of chromosome 7) and probes in black are normal for the other chromosomes studied in these kits. (B) SNP-A (CytoScanHD Array; Affymetrix, Santa Clara, USA): Diagrams generated by ChAS (Affymetrix) for chromosome 2 (above) and chromosome 7 (below). In “Copy number state”, line in 2.00 indicates a normal copy number (diploid), line in 1.00 indicates a deletion and line in 3.00 indicates a duplication/gain. Lines with intermediate values between 1.00 and 2.00; 2.00 and 3.00 indicate mosaicism. In “Allele Difference”, three lines indicate a normal genotype (AA, AB and BB alleles), two lines indicate deletion (A and B alleles) or loss of heterozygosity (AA and BB alleles) and four lines indicate duplication (AAA, AAB, ABB and BBB alleles). SNP-A revealed the result arr[hg19] 2p25.3p16.2(12770_54276429)x3[0.8],2q35q37.3 (219576639_242783384)x1,(7)x1 where there was a partial duplication of the short arm of chromosome 2 (A and C), presenting clonal mosaic data of the duplicated region in 80% of the cells, and partial deletion of the long arm of chromosome 2 (B and D). This result can explain add (2) (q35) present in the karyotype, indicating the origin of the additional material. In addition, the monosomy of chromosome 7 was confirmed.](https://static.elsevier.es/multimedia/15168484/0000003900000004/v2_201911281025/S1516848417300920/v2_201911281025/en/main.assets/thumbnail/gr2.jpeg?xkr=ue/ImdikoIMrsJoerZ+w93OM6WmS6o9DeZl+SVh74uo=)





