Prevalence of RhD negative phenotype in Nigeria is low; this leads to scarcity of RhD negative red cells for transfusion. Serological and molecular genotyping of RhD negative individuals for weak D types could reduce this scarcity. The aim of this study was to determine the serological prevalence and molecular types of weak D phenotypes among blood donors and pregnant women in Kano, Nigeria.
MethodsA total of 4482 blood donors and pregnant women from three hospitals in Kano were recruited. An indirect antiglobulin test was used to determine weak D phenotypes. Molecular genotyping was performed on genomic DNA from whole blood amplified by polymerase chain reaction sequence-specific primers (PCR-SSP) with agarose gel electrophoresis.
ResultsThe mean age of the participants was 26.50 ± 5.79 years. The prevalence of the RhD negative phenotype was 4.2% (189/4482). Of the 189 RhD negative phenotypes, 20 (10.6%) were weak D positive. Molecular genotyping of the 20 Weak D positive phenotypes revealed 15 (75%) weak D type 4, of which 11 were due to the RHD*09.03 and RHD*DAR3 (T201R, F223V) polymorphisms and 4, due to RHD* 08.01 and RHD* DFV polymorphisms; 2 (10%) were due to the 602 C>G polymorphism, while the remaining 3 (15%) constituted partial D or other rare weak D types.
ConclusionThe prevalence of weak D positive phenotypes is high in this study; weak D type 4 is the most common RhD genetic variant. Routine serologic weak D testing of RhD negative blood and molecular genotyping should be encouraged in resource-limited settings.
The Rh blood group is clinically second only to the ABO in relevance to transfusion medicine.1,2 The system is made up of 5 antigens (D, C, c, E and e) that are encoded by two closely linked homologous genes, RHD and RHCE, located on the short arm of chromosome 1.3 The RHD gene encodes the RhD antigen, while the RHCE gene encodes RhCE antigens. Clinically, the most important Rh antigen is D because of its high propensity for immunogenicity.1 The RhD polypeptide is made up of 417 amino acid sequences that are folded into a 12 segments of the trans-red cell membrane protein with 6 extracellular/intracellular loops.3 Amino acid substitution (s) involving the extracellular, transmembrane, or intracellular domains of the protein, could lead to qualitative or quantitative under-expression of the RhD antigen that is serologically typed as the RhD negative phenotype.2,3 This substitution results in production of RhD variants that are classified into: partial D, weak D, DEL and non-functional alleles.3 Weak D phenotypes develop as a result of substitution in the transmembrane or intracellular domains of the RhD protein, with consequent quantitative under-expression of the protein.3 Depending on the type or subtype, weak D positive individuals can face or pose significant challenges in transfusion medicine, with associated clinical sequelae.
In Nigeria, routine Rh typing in transfusion medicine involves serological typing solely for the RhD antigen; participants in whom the RhD antigen is present are considered RhD positive, while those without the antigen are termed RhD negative. While individuals who are RhD positive do not face or pose Rh-related problems in transfusion medicine, those who are RhD negative constitute an important source of concern to both clinicians and blood bank staff, either as potential recipients or as donors of red cells for transfusion. As potential recipients of red cell transfusion, RhD negative individuals can only be transfused safely with RhD negative red cells, while as potential donors, some individuals may have a weak D variant that is capable of sensitizing a given recipient.1,2
The prevalence of the RhD negative phenotype in Nigeria is low, being at 5.1%.4 The low prevalence means reduced availability of RhD negative red cells for transfusion to RhD negative individuals at critical times of need. This scarcity puts further constraints on the already inadequate transfusion services in the country. Furthermore, in RhD negative pregnant women, alloimmunization can lead to hemolytic disease of the fetus and newborn in subsequent pregnancies with RhD positive fetuses. Even though preventable with prophylactic administration of anti-D immunoglobulin, the cost and availability of the drug in low-income countries can sometimes be prohibitive. Serological typing of RhD negative individuals for the presence of the weak D type and subsequent molecular genotyping of weak D positive individuals could reduce the scarcity of RhD negative red cells by ensuring that weak D positive individuals are transfused with RhD positive red cells, thus conserving RhD negative red cells for individuals that are truly RhD negative.5 It will also prevent the unnecessary and costly administration of anti-D immunoglobulin to weak D positive women after the delivery of RhD positive babies.5 While a number of studies have explored the prevalence of serological weak D phenotypes in Nigeria,6–8 to our knowledge, none performed molecular genotyping of the weak D phenotypes.
The objective of this study was to determine the serological prevalence and molecular variants of weak D phenotypes among RhD negative blood donors and pregnant women in Kano, Nigeria.
Methods and materialsStudy area, design and selection of participantsThe study was a descriptive cross-sectional study conducted in Kano, northwest Nigeria, between September 2019 and February 2020. The convenience sampling technique was used to recruit participants. Female participants were recruited from the antenatal clinics of the Aminu Kano Teaching Hospital, Murtala Muhammad Specialist Hospital and Muhammad Abdullahi Wase Teaching Hospital, while male participants were blood donors at the donor clinics of the three hospitals. A total of four thousand four hundred and eighty-two (4482) participants, consisting of 3609 males and 873 females, were recruited. The mean age of the participants was 26.50 ± 5.79 years. Of the 4482 participants who took part in the study, 3652 (81.48%) were blood donors, while 830 (18.52%) were pregnant women.
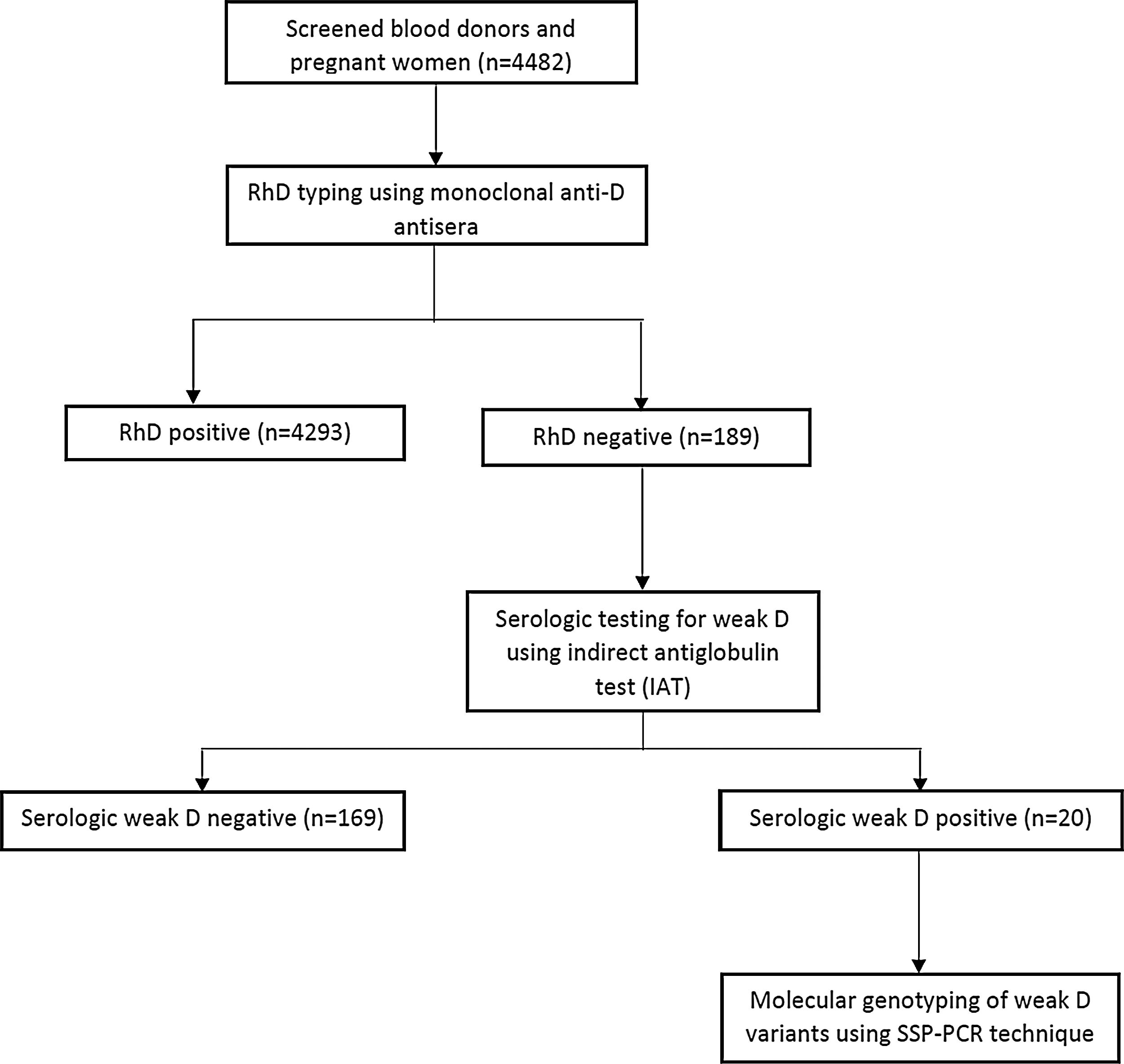
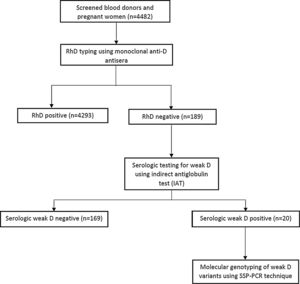
Serological typing for RhD and weak D phenotypesEight milliliters (8 mL) of venous blood was aseptically drawn from each participant and 5 mL transferred into an ethylenediaminetetraacetic acid (EDTA) container and the remaining 3 mL was dispensed into a plain tube and allowed to coagulate at room temperature. The clotted samples were centrifuged at 1000 g for 5 min, after which red cells were separated from serum using a plastic Pasteur pipette. The red cells were washed three times in saline and a 5% saline suspension of red blood cells was made. Rh blood group phenotypes were determined using commercial monoclonal anti-D reagents (Plamatec Lab., Ltd., Bridport, UK), performing the conventional tube technique, as described by Regan.9 Samples that had RhD negative or weak (≤2+) agglutinations were then tested for the weak D phenotype using the indirect antiglobulin test (IAT), as described below. Figure 1 shows participant selection and the testing algorithm.
Weak D typing was performed by the IAT, as described by Regan.9 Briefly, all tubes that showed no or weak (≤2+) agglutination with the initial anti-D serum in the RhD typing were incubated at 37 °C for 60 min and then washed three times in saline. The red cells were then mixed with two drops of anti-human globulin (Plamatec Lab., Ltd., Bridport, UK) and centrifuged at 1000 g for 2 min. The contents were examined for agglutination macroscopically and microscopically; samples that showed moderate to strong agglutination were regarded as weak D positive. Hence, we define serological weak D phenotype here as blood samples that show no or weak (≤2+) agglutination conventional anti-D antiserum, but moderate to strong (more than 2+) agglutination in the IAT using a potent monoclonal anti-D reagent.2
Molecular genotyping of weak D phenotypesDNA extractionGenomic DNA was extracted from whole blood using the QIAamp DNA Blood Mini Kit (Qiagen, Hilden Geschäftsführer, Germany), as described by Gassner et al.10
Sequence-specific primer PCR amplification of extracted DNAThe DNA analysis of the weak D positive phenotypes was performed by the PCR-SSP RHD exon scanning, as previously described.10 Sequence specific primers for the detection of weak D types were selected based on a previous study.11 The primer sequences used are presented in Table 1.
Primers used for the PCR-SSP for weak D types.
| Primer name | Exon | Nucleotide exchange | Nucleotide Sequence | Specificity | Size(kb) |
|---|---|---|---|---|---|
| Gwd4 /F | Exon 4 | 602 C>G | 5′ AGACTACCACATGAACATGATGCACA 3′ | Weak D type 4 (T201R) | 138 |
| Gwd4b /R (wild type allele) | 3′ CAGACAAACTGGGTATCGTTGCTG 5′ | ||||
| Gwd4 /F | Exon 4 | 602 C>G | 5′ AGACTACCACATGAACATGATGCACA 3′ | Weak D type 4 (T201R) | 138 |
| Gwd4b /R | 3′ CAGACAAACTGGGTATCGTTGCTC 5′ | ||||
| Rh223vf/F (wild type allele) | Exon 5 | 667 T>G | 5′ TTGTGGATGTTCTGGCCAAGTT 3′ | Weak D type 4 (F223V) | 168 |
| Ga51/R | 3′ CTGCTCACCTTGCTGATCTTCCC 5′ | ||||
| Rh223vf/F | Exon 5 | 667 T>G | 5′ TTGTGGATGTTCTGGCCAAGTG 3′ | Weak D type 4 (F223V) | 168 |
| Ga51/R | 3′ CTGCTCACCTTGCTGATCTTCCC 5′ | ||||
| HGH-F | HGH gene | 5′ TGCCTTCCCAACCATTCCCTTA 3′ | 434 | ||
| HGH-R | 3′ CCACTCACGGATTTCTGTTGTGTTTC 5′ |
The PCR was performed in a Gene touch thermal cycler (TCE 48 FA, China). A total PCR reaction volume of 15.0 μL was prepared for each reaction. Each reaction volume was comprised of 1.50 μL 10 × PCR buffer, 0.51 μL of each 10 pM primer, 0.75 μL of MgCl and 1.0 μL of DNA. Each PCR reaction was mixed thoroughly. The PCR reaction conditions were an initial denaturation at 95 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 65 °C for 30 s and extension at 72 °C for 1 min and then a final extension at 72 °C for 10 min, with the product being held at 4 °C. A primer for the human growth hormone was used as an internal control.
Visualization of amplified productFollowing the amplification, 4 µL of the amplified DNA was added to 2 µL of bromophenol blue DNA loading dye and electrophoresed in 2% agarose gel stained with 4 µL of ethidium bromide at 110 V for 30 min. The DNA bands were visualized and documented using a UV transilluminator and the size of the amplified product was determined by comparison against a 100 bp DNA ladder.
Statistical analysisThe data were analyzed on the IBM SPSS version 23.0 (IBM, Armonk, New York, USA).
Ethics considerationThe ethics approval was obtained from the Research Ethics Committees of the Aminu Kano Teaching Hospital and Kano State Ministry of Health, Nigeria. Written informed consent was obtained from all participants. The study complied with the ethics rules stated in the Declaration of Helsinki.
ResultsOf the 4482 blood samples typed for RhD, 189 (4.2%) samples were RhD negative, while 4293 (95.8%) were RhD positive. Of the 3652 blood donors, 155 (4.2%) were RhD negative, while 34 (4.1%) of the 830 pregnant women, were RHD negative.
Of the 189 RhD negative samples (155 blood donors and 34 pregnant women), 20 (10.6%) were weak D positive. Of the 155 RhD negative samples among blood donors, 15 (9.7%) were weak D positive. Similarly, of the 34 RhD negative samples among pregnant women, 5 (14.7%) were weak D positive phenotypes, as shown in Table 2.
Molecular genotyping of the 20 weak D positive samples revealed that 15 (75%) were weak D type 4, of which 11 were due to 602 C>G;667 T>G (T201R, F223V) polymorphism (exons 4 and 5), while 4 were due to 667 T>G polymorphism (exon 5); two (10%) were due to single polymorphism (exon 4) for 602 C>G (Eurasian D cluster, weak D type 40), while the remaining three (15%) samples constituted ‘other’ types, probably partial D or other rare weak D types, as shown in Table 3.
Molecular genotyping of the Weak D positive participants.
| Cluster | Alteration | ISBTa Name | Frequency (N) | Proportion (%) |
|---|---|---|---|---|
| Weak D type 4 cluster | ||||
| Weak D type 4.0.1 | 602 C>G;667 T>G | RHD*09.03, | 11 | 55 |
| RHD* DAR3 | ||||
| Partial D | 667 T>G | RHD* 08.01 | 4 | 20 |
| RHD* DFV | ||||
| Weak D type 40 | 602 C>G | RHD*01W.40 | 2 | 10 |
| Eurasian D cluster | RHD*weak D 40 | |||
| Partial or other Weak D variants | – | – | 3 | 15 |
This study provides an insight into the burden and significance of serological weak D phenotypes and RhD genetic variants in a resource-limited setting. The prevalence of the RhD negative phenotype among the participants in this study was low, being 4.2%; this is similar to findings of other studies in Nigeria and in many countries in Africa that reported prevalence rates ranging from 2.0 to 5.1%.4,12–15 This prevalence is higher than that found in people of Asian ancestry, for whom the figure of 1% or less has been reported,16 but much lower than findings in European and North American populations, with prevalences of the RhD negative phenotype ranging from 15 to 17.3%.16,17
The prevalence of the serologic weak D positive phenotype in this study was 10.6%. This prevalence is higher than the prevalences reported from different parts of Nigeria, ranging from 0.95 to 4.9%7,8,18 and other African countries, registering 2.0–2.6%.12,19,20 A moderately high prevalence of D variants has been reported among African-Americans in the USA (9.8%),21 as well as in Canada (36%)22 and Brazil (8.4%).23 Ethnic and racial differences in the expression of RhD and its variants have been widely reported. The State of Kano is the commercial center of northern Nigeria, boasting many of Nigeria’s ethnic nationalities; this could explain the relatively high prevalence found in this study. Other factors that could influence the prevalence of the serological weak D phenotype are the type and sensitivity of the laboratory technique used for the weak D testing.2 In this study, we used the conventional tube method to determine the prevalence of the serological weak D phenotype. The use of higher sensitivity techniques, such as the automated gel column or the solid-phase technology coupled with a blend of potent recombinant monoclonal anti-D antisera, has been reported to increase the prevalence of the weak D phenotype.2
The weak D type 4 was the most common genetic variant in this study, with the weak D subtype 4.0.1 as the most frequent subtype. Various studies have reported the weak D type 4 as the most common type in Africans and people of African ancestry.23–25 However, in white populations, the less immunogenic weak D types 1, 2 and 3 are the most prevalent.22 While some pregnant women with weak D types 1, 2 and 3 do not form alloantibodies, the current recommendation being that such women can safely be transfused with RhD positive blood, some subtypes of weak D type 4 are found to be immunogenic.9 Hence, transfusion recommendations in patients with weak D type 4 variants require more circumspection to avoid allosensitization.2,26 The relatively high frequency of the weak D type 4 in this study means that transfusing weak D positive recipients with RhD positive red cells should be undertaken with caution in this environment to avoid potential alloimmunization and molecular genotyping of the RHD gene should be encouraged to determine the exact RhD genotype for transfusion decision-making under these circumstances. We recommend that this preponderance of the potentially immunogenic weak D type 4.0.1 in our population be taken into consideration when developing clinical guidelines for the transfusion of pregnant women with the serologic weak D phenotype. For example, if a pregnant woman or a patient is found to have a weak D phenotype, based on the serology in a resource-constrained setting with a high prevalence of the immunogenic variants, such as the weak D type 4 found in this study, and where further genetic analysis is not feasible, it would be a wise and safe recommendation to transfuse such individuals solely with RhD negative red cells. Such practice will help avoid the development of potential adverse transfusion outcomes, such as allosensitization, delayed hemolytic transfusion reactions or hemolytic disease of the fetus and newborn in subsequent pregnancies. If such practice is implemented, RhD negative pregnant women with serologic weak D phenotypes would be regarded as candidates for anti-D immunoglobulin prophylaxis.2 Our study did not find the DEL genotype among the study participants, this being consistent with the findings in African-Americans.2 In contrast to this, the DEL genotype has a frequency of 17–30% and 0.1% among RhD-negative Asian and white populations, respectively.
In this study, we used the IAT to determine serologic weak D phenotypes. Although this technique can identify individuals with RhD variants, it lacks the capacity to categorize them into different weak D types and subtypes, partial D and other rare variants. This categorization is sometimes necessary because not all RhD variants develop alloantibodies.22 Molecular genotyping, on the other hand, can identify the different RhD variants. Moreover, while the IAT for serological weak D testing is relatively inexpensive and easy to perform, the same cannot be said of routine RhD genotyping, especially in a resource-limited setting. Genotyping of the RHD gene can, however, be cost-effective when the cost of anti-D immunoprophylaxis in pregnant women is considered, especially in ethnic groups in which the prevalence of immunogenic variants is prevalent.27 Given the intrinsic weakness of national blood transfusion services in low- and middle-income countries (LMICs), most of these countries operate hospital-based blood services with routine immunohematological testing conducted in in-hospital laboratories. To further optimize potential cost-savings for genotyping under these circumstances, we recommend a centralized model of RHD genotyping to resolve serological weak D phenotypes, in which more rigorously controlled molecular testing can be conducted in designated national or regional reference laboratories. This could ensure cost-saving through economy of scale, in addition to potentially higher quality testing.
Altogether, the implication of our findings on the safety and availability of allogenic blood for transfusion in resource-limited settings is at least two-fold: 1) Given the high prevalence of the serological weak D phenotype found in this study, the lack of routine testing for this phenotype in most clinical settings at our hospitals means that a significant proportion of serologically-typed RhD negative persons with the serological weak D phenotype are missed. This could lead to avoidable alloimmunization if RhD negative recipients are transfused with red cells that are weak D positive. Additionally, RhD negative pregnant women with the weak D phenotype might be unnecessarily administered anti-D immunoglobulin, with the attendant high cost of the medication. Considering that healthcare expenditure in LMICs, such as Nigeria, is largely out-of-pocket and this places a significant financial burden on families. 2) In the setting of weak health systems and scarce allogenic blood for transfusion, as found in LMICs, significant time and financial and logistical resources can be wasted scouting for RhD negative blood units when potential recipients are actually RhD positive with the weak D phenotypes. These resources can be saved and channeled to other uses when the exact phenotype is known.
ConclusionThis study found a higher prevalence of serological weak D in a Nigerian population than had previously been reported. For the first time in this population, we performed molecular genotyping of the RHD gene in weak D positive phenotypes and found weak D type 4 subtype 4.0.1 as the most prevalent molecular variant. Molecular genotyping of the RHD gene should be encouraged in blood donors, pregnant women and transfusion recipients of RhD negative, weak D positive red cell phenotypes, to avoid potential allosensitization and wastage of scarce allogenic blood in resource-limited settings.
Contribution of the authorM. U. D., Y.A.A., and S. A. I. conceptualized the study. MUD recruited participants and performed laboratory analyses. M. I. G. analyzed the data and wrote the initial draft of the manuscript. All authors critically reviewed, approved the final version and contributed to the intellectual content of the manuscript.
Conflicts of interestThe authors declare no conflicts of interest.