T-cell prolymphocytic leukemia (T-PLL) is a rare hematologic neoplasm that accounts for less than 2% of mature malignant lymphoid leukemia.1 The T-PLL is usually aggressive, with hepatosplenomegaly, generalized lymphadenopathy, lymphocytosis usually higher than 100 × 109/L and very poor clinical outcomes.2 It affects older adults with a mean age of 65 years, with a male predominance. Most cases of T-PLL are acquired, but some cases evolve from constitutional diseases, such as ataxia-telangiectasia, caused by the ATM gene mutation (ataxia-telangiectasia mutant).3 In most cases, the cell morphology is characterized by a predominance of small to medium lymphoid cells with basophilic cytoplasm and round, oval, or irregular nuclei with prominent nucleoli and condensed chromatin.4 T-PLL cells are post-thymic cells, positive for markers of mature T cells and are negative for the TdT and CD1a . The cells are generally positive for the CD2, CD3 and CD5 and are strongly positive for the CD7. They also express the cyTCL1+ (> 90%). Most cases express the CD4 (65%), few cases show the CD8+ (13%) and co-expression of the CD4 and CD8 occurs in 21% of the cases.5 An analysis of 24 variable regions of the TCR Vβ2 repertoires by flow cytometry (Vbeta) can be used to confirm clonal alpha-beta T lymphocytes and is characteristically performed on peripheral blood. This technology may help confirm the T-cell clonality in the T-cell prolymphocytic leukemia.6
In the conventional cytogenetic analysis, the T-PLL is characterized by a complex karyotype (CK) involving mainly chromosomes 14, 8, 11 and X. The inversion of chromosome 14, reported as the inv(14)(q11;q32), is the most common abnormality observed (80% of the cases) and the tandem translocation t(14;14)(q11;q32) occurs in 10% of the patients.7 The gene encoding the T-cell receptors alpha (TRA) and delta (TRD) are located on the 14q11.2 and the TCL1 (T-cell leukemia/lymphoma1) proto-oncogene is located on the 14q32. These two rearrangements cause the juxtaposition of the TCL1 and TCR genes, activating the expression of the TCL-1.1
The t(X;14)(q28;q11) translocation observed in approximately 20% of the cases involves the MTCP1 gene located in the X(q28). The TCL-1 and MTCP1 can induce the T-PLL.1 Chromosomal abnormalities involving the long and short arm of the chromosome 8, or trisomy, are seen in 70 to 80% of the cases.5 The long arm isochromosome or trisomy of chromosome 8 results in additional copies of the MYC gene, located on the 8q24.2. Other recurrent chromosomal abnormalities observed in the T-PLL are deletions on the long arm of chromosome 11, del(11)(q22.3), where the ATM gene is located, and deletions or rearrangements on the short arm of chromosome 17, where the TP53 gene is located.1
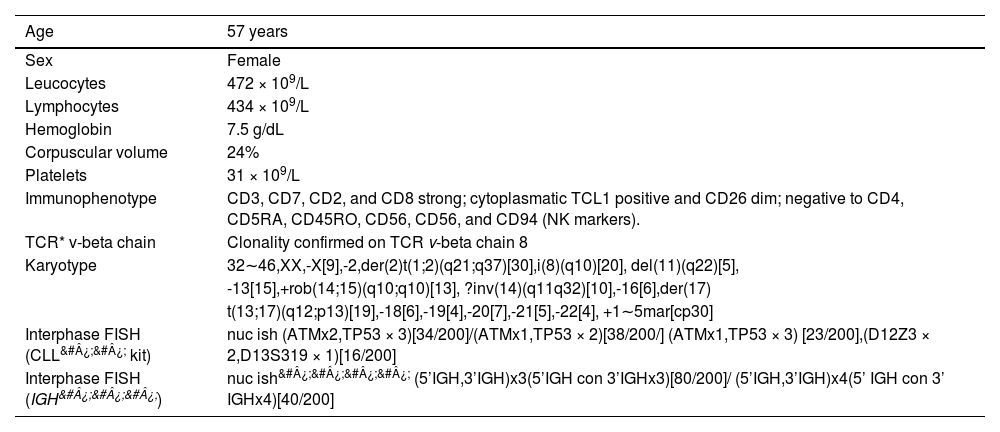
CLINICAL PRESENTATIONA 57-year-old woman, previously diabetic, multipara, presented with abdominal pain and distension, weight loss, asthenia and a decreased general condition for two months. The examination revealed a splenomegaly (19 cm RCE), hepatomegaly (18 cm RCD) and generalized lymphadenopathies of no more than 2 cm. The hemogram showed anemia (Hb 7.5 g/dL), significant leukocytosis (472 × 109/L) with 92% of lymphocytes, of which more than 50% were prolymphocytes, and a platelet count of 31 × 109/L.
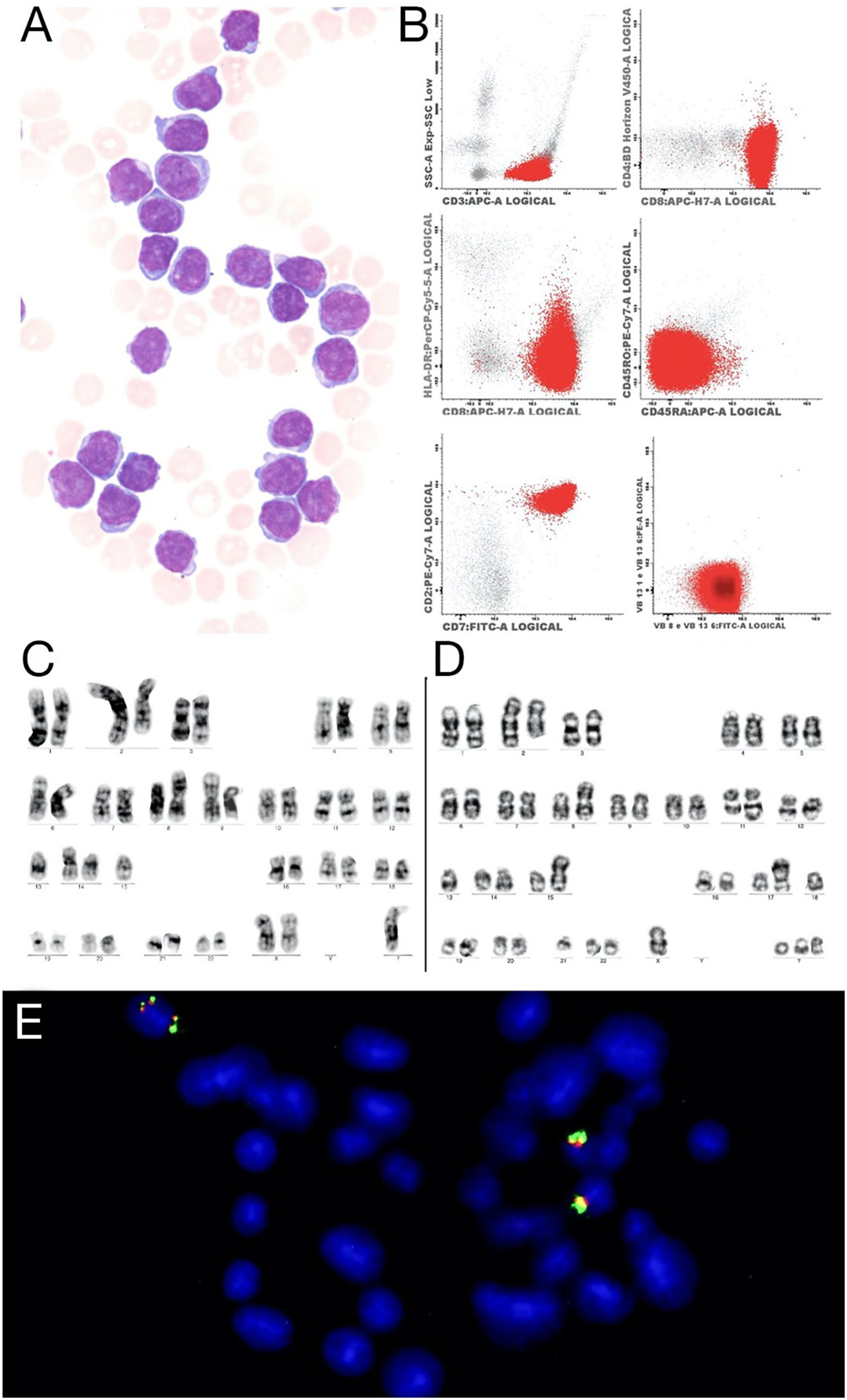
The multiparametric flow cytometry analysis (MFC) in peripheral blood showed a mature T cell lineage strongly positive for the CD3, CD8, CD7 and CD26, negative for the CD4, CD45 RA, and RO and NK marker isoforms (CD56, CD57, CD94), but positive for the cytoplasmic TCL1. The clonality was confirmed in the abnormal CD8 cells by the expressed TCR Vβ2 repertoires (Figure 1 A). The conventional cytogenetic examination showed a complex karyotype. The patient was treated with chemotherapy (fluorouracil, cyclophosphamide and mitoxantrone), with no response (lymphocytes low at 100 × 109/L). As she continued to present lymphocytosis, lymphadenomegaly and hepatosplenomegaly) she was treated with alemtuzumab (anti-CD52), with the expectation of completing 12 cycles, but showed progression within three months, requiring discontinuation of the medication. The patient was referred for palliative care.
(A) Prolymphocytes in peripheral blood, Wright-Giemsa 100x (B) Flow cytometry showed strongly positive for CD3, CD8 and CD7. (C) Metaphase presenting a complex karyotype, with a der(2)t(1;2), i(8)(q10), der(9), der(14) and a marker chromosome. (D) Metaphase shows complex karyotype with a der(2)t(1;2), i(8)(q10), der(11q) inversion of chromosome 14,rob(14;15)(q10;q10), t(13;17) and three marker chromosomes. (E) FISH with the LSI IGH Dual Color, Break Apart Rearrangement Probe (Vysis) showed four signals of the IGH gene, two of them show on the same chromosome.
The morphological evaluation was performed on the peripheral blood smear stained with Wright-Giemsa. The conventional cytogenetic analysis was performed on heparinized peripheral blood cells cultivated in the RPMI 1640 medium with phytohemagglutinin for 72 hours at 37°C at diagnosis. The chromosome analysis was performed by GTG-banding and thirty metaphases were analyzed. The interphase and metaphase FISH (Fluorescence in Situ Hybridization) were performed on fixed cells from peripheral blood cultures. The panel utilized was the Vysis CLL FISH probe (Vysis: REF: 04N02-022; Abbott Molecular: Illinois, USA), with four probes: LSI ATM (11q22.3) spectrum green, LSI TP53 (17p13.1), spectrum red, CEP12 (12p11.1-q11) spectrum green, LSI D13S319 (13q14.3) spectrum red and LSI 13q34, spectrum aqua. Besides this panel, the LSI IGH Dual Color, Break Apart Rearrangement Probe (Vysis: REF: 08L63-020 Abbott Molecular: Illinois, EUA), LSI MYC Break Apart Probe (Cytocell: REF:LPH010), AML1::ETO (RUNX1::RUNX1T1) Translocation, Dual Fusion (Cytocell:REF: CE-LPH 026, and the PML::RARA, Translocation, Dual Fusion (Cytocell: REF: LPH023; Oxford, UK) were also applied. The FISH protocol was performed according to the manufacturer's instructions. The conventional cytogenetic and the interphase FISH results were reported according to The International System for Human Cytogenetic Nomenclature (ISCN), 2016. The multiparametric flow cytometry was performed with an 8-color panel and stain-then-lyse protocols were used to analyze the expression of cell surface markers. Leukocytes were stained with antibodies against the follow antibodies, fluorescence and clones: CD2 PeCy7 (L303-1), CD3 V450 (UTH1), CD4 PercPcy5.5 (SK3), CD5 PercpCy5.5 (L17F12), CD7 FITC (4H9), CD8 APCH7 (SK1), CD19 Pecy7 (SJ25C1), CD26 PE (L272), CD27 FITC (L128), CD28 APC (28.2), CD45 V500C (clone 2D1), CD56 PE (N901), CD57 FITC (HNK1), CD94 APC (HP-3D9), TCR alpha-beta FITC (WT31) and TCR gamma-delta PE (11F2), cytoplasmatic TCL1 APC (Ebio1-21), Granzyme PE (CLB-GB11) and Perforin FITC (GG9).
Normal lymphoid cells within the specimens served as internal controls to define positive and negative staining. The bright immunofluorescence intensity was defined as higher than that observed in normal T cells and the dim staining was defined as lower than that observed in normal T cells.
The TCR Vβ2 repertoire was evaluated using the IOTest Beta Mark TCR Vbeta Repertoire kit (Beckman Coulter) that has antibodies against 24 variable regions of the T-cell-receptor beta chain: Vβ 1, Vβ 2, Vβ 3, Vβ 4, Vβ 5.1, Vβ 5.2, Vβ 5.3, Vβ 7.1, Vβ 7.2, Vβ 8, Vβ 9, Vβ 11, Vβ 12, Vβ 13.1, Vβ 13.2, Vβ 13.6, Vβ 14, Vβ 16, Vβ 17, Vβ 18, Vβ 20, Vβ 21.3, Vβ 22 and Vβ 23. These antibodies, labeled with FITC, PE, or FITC and PE, were placed in 8 test tubes with antibodies that separated the T lymphocytes with an aberrant immunophenotype from normal leukocytes that served as internal polytypic controls for the Vbeta analysis. The discriminating antibodies included the CD3 APC, CD8 APC-H7 and CD4 V450.
Quantification and characterization of leucocytes and pathological cells were performed using the FACSCanto II® cytometer and Infinicyt™ software (Cytognos, Salamanca, Spain — version 1.8).
RESULTSIn this study, we report the case of a patient with a rare CD8 T-prolymphocytic leukemia presenting with an elevated peripheral blood white blood cell count and a very complex karyotype with a rare derivative chromosome der(2)t(1;2)(q21;q37), abnormal immunophenotype (Figure 1A), and progressive disease, despite treatment with alemtuzumab. The clinical and laboratory data of the patient are summarized in Table 1.
Clinical and laboratory data of the patient
TCR*: T cell receptor; CLL&#¿;&#¿;: Chronic lymphocytic leukemia;
IGH&#¿;&#¿;&#¿;: Immunoglobulin Heavy Chain; nuc ish&#¿;&#¿;&#¿;&#¿;: interphase/ nuclear in situ hybridization
The conventional cytogenetics analysis showed a complex karyotype with more than five chromosomal abnormalities. The number of chromosomes varied from 32 to 46. The abnormalities present were: an unbalanced translocation involving the long arms of chromosomes 1 and 2, producing a der(2)t(1;2)(q21;q37) and resulting in a partial trisomy of one segment of the long arm of chromosome 1, observed in all metaphases analyzed (n = 30); an isochromosome for the entire long arm of one chromosome 8, i(8)(q10); a derivative of chromosome 14, the result of inv(14)(q11q32) or t(14;14)(q11;q32); a Robertsonian translocation, rob(14;15)(q10;q10); a derivative chromosome 17 originating from the translocation t(13;17)(q12;p13); a deletion on the long arm of chromosome 11, del(11)(q22.3), and; many marker chromosomes (see the complete karyotype in Table 1). Some of these abnormalities were confirmed by the metaphase FISH. The interphase FISH with the CLL FISH probe showed the del(11)(q22.3), an extra signal of the TP53 gene and the del(13)(q14.3). The interphase FISH with the IGH break-apart probe showed one and two additional copies of the variable segment of this gene (FISH results in Table 1). This probe was also used on metaphases that show four IGH gene signals (Figure 1 D). Conventional cytogenetics shows an inversion of chromosome 14 and a marker chromosome (Figure 1 B-C), which resembles a derivative chromosome 14. The isochromosome for the entire long arm of one chromosome 8 was confirmed by the LSI MYC break-apart probe and by the probe AML1::ETO (RUNX1::RUNX1T1) (not shown), resulting in trisomy of the long arm of chromosome 8 and, consequently, a gain of one copy of the MYC gene.
The derivative of chromosome 17 is the result of a translocation t(13;17)(q12;p13), which was confirmed by the probes LSI 13q34 (aqua) and D13S319 (orange), both for the long arm of chromosome 13, without the losses of these loci. A separate hybridization with the LSI TP53 probe also confirmed this rearrangement. The abnormality observed in chromosome 15 was interpreted as a Robertsonian translocation with chromosome 14, confirmed by the metaphase FISH (not shown).
DISCUSSIONThe present case was diagnosed through peripheral blood lymphocyte morphology, clinical evaluation and imaging examinations. The immunophenotype, conventional cytogenetics analysis and interphase/ metaphase FISH confirmed the diagnosis of T-PLL, according to the recommendations established by the T-PLL International Study Group (TPLL-ISG). Our case shows a CD8 positive T-PLL and a complex karyotype with more than five chromosome abnormalities. The der(2)t(1;2)(q21;q37) observed in all metaphases is a rare anomaly in hematological malignancies with unknown clinical significance. This abnormality has not yet been described in T-PLL, but it has been described in 18 cases of other hematologic diseases, such as acute myeloid leukemia (AML) and multiple myeloma (MM), and, less frequently, in lymphoid malignancies, reported in in the Atlas of Genetics and Cytogenetics in Oncology and Hematology. This derivative is also described in hepatoblastoma as the most common structural abnormality observed (18% of the cases).8
The second most frequent chromosome abnormality observed was the isochromosome 8, producing an additional copy of the MYC gene. The third most frequent abnormality observed was the der(17)t(13;17), confirmed by the metaphase FISH. The interphase FISH showed three copies of the TP53 gene. The fourth most observed chromosome abnormality was a Robertsonian translocation involving the acrocentric chromosomes 14 and 15. This abnormality was described in two cases of CLL.9,10 One case of prolymphocytic leukemia with a Robertsonian translocation t(13;15)(q11;p12) was reported by Morgan et al., (1987).11 The present case reported a patient with seven children, which rules out the possibility of the Robertsonian translocation being constitutional. The inversion of chromosome 14 and/or the translocation t(14;14) was observed in ten metaphases. This rearrangement is common in T-PLL and is reported in 64% of the cases.7 The overall survival (OS) of patients with 14q abnormalities is similar to those without these abnormalities.12 The interphase FISH showed the presence of 3 to 4 copies of the IGH gene, located on 14q32.3, confirmed by the metaphase FISH, which had shown two signals on the same chromosome, reinforcing the rearrangement involving two chromosomes 14. The cytogenetics analysis also showed a deletion in 11q, confirmed by the interphase FISH with the probe for the ATM gene. This alteration is also described in a case report of a female patient with T-PLL who had a CK with more than five chromosome abnormalities.13 Many chromosomal losses were observed, such as the losses of X, 18, 19, 20, 21 and 22. These missing chromosomes are probably involved in forming the different marker chromosomes present. The involvement of the chromosomes X, 8, 14 and 11 have already been described in 21 T-PLL cases and most of these abnormalities were detected more by the FISH than by cytogenetics.7 Yang et al., (2021) have found similar abnormalities in a 68-year-old male T-PLL patient with significant leukocytosis and a history of T-cell lymphoproliferative disorder. In this case, the cytogenetic analysis uncovered a complex karyotype (CK), including the dual translocation of chromosome 14, rearrangements on chromosomes 9p and 5p, isochromosome 8, deletion 11q, and monosomy 17. The FISH analysis revealed TRD (14q11.2) rearrangements, a loss of ATM and CDKN2A signals and gains in RELN, TES and MYC signals. 14
The patients with a CK had a poorer OS and no individual cytogenetic abnormality impacted the OS. The presence of more than five chromosome abnormalities contributes to a decrease in survival, with an OS of less than 11 months versus 22 months in patients with ≤ 5 chromosome abnormalities.12 The complete laboratory analysis, including conventional and molecular cytogenetics analysis, is fundamental for the T-PLL evaluation, prognosis and therapeutic intervention. Also, it is important to emphasize that this method is accessible to many health centers and cheaper than molecular methods for places without this technology.
We would like to thank my colleagues from Hospital de Clínicas de Curitiba, Alexandra Cristina Senegaglia, Ana Teresa Schmid-Braz, Denise Cristine Coutinho, Douglas Câmara de Oliveira, Lismeri Wuicik Merfort, Miriam Lacerda Barbosa, Rodrigo Miguel Bendlin and Tamara Borgonovo, for their contributions to this paper.
- Início
- Todos os conteúdos
- Publique seu artigo
- Sobre a revista
- Métricas