Chronic myeloid leukemia is a myeloproliferative disorder characterized by the Philadelphia chromosome or t(9;22)(q34.1;q11.2), resulting in the break-point cluster region-Abelson tyrosine kinase fusion gene, which encodes a constitutively active tyrosine kinase protein. The Philadelphia chromosome is detected by karyotyping in around 90% of chronic myeloid leukemia patients, but 5–10% may have variant types. Variant Philadelphia chromosomes are characterized by the involvement of another chromosome in addition to chromosome 9 or 22. It can be a simple type of variant when one other chromosome is involved, or complex, in which two or more chromosomes take part in the translocation. Few studies have reported the incidence of variant Philadelphia chromosomes or the breakpoints involved among Brazilian chronic myeloid leukemia patients.
ObjectiveThe aim of this report is to describe the diversity of the variant Philadelphia chromosomes found and highlight some interesting breakpoint candidates for further studies.
Methodsthe Cytogenetics Section Database was searched for all cases with diagnoses of chronic myeloid leukemia during a 12-year period and all the variant Philadelphia chromosomes were listed.
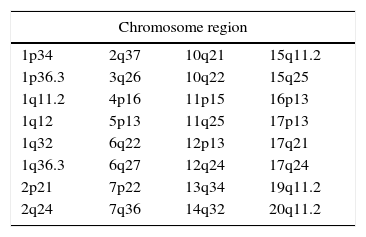
ResultsFifty (5.17%) cases out of 1071 Philadelphia-positive chronic myeloid leukemia were variants. The most frequently involved chromosome was 17, followed by chromosomes: 1, 20, 6, 11, 2, 10, 12 and 15.
ConclusionAmong all the breakpoints seen in this survey, six had previously been described: 11p15, 14q32, 15q11.2, 16p13.1, 17p13 and 17q21. The fact that some regions get more frequently involved in such rare rearrangements calls attention to possible predisposition that should be further studied. Nevertheless, the pathological implication of these variants remains unclear.
Chronic myeloid leukemia (CML) is a myeloproliferative disorder characterized by the presence of the Philadelphia chromosome (Ph) which is the derivative chromosome 22 of the translocation t(9;22)(q34.1;q11.2). Due to this rearrangement, the break-point cluster region (BCR) gene at position 22q11.2 is juxtaposed to the c-Abelson (ABL1) gene at 9q34, resulting in the BCR-ABL1 fusion gene, encoding a constitutively active tyrosine kinase protein. The identification of this abnormality is important for the diagnosis of the disease as determined by the WHO Tumor Classification1 and for treatment purposes. The first therapeutic choice, tyrosine kinase inhibitors, has shown great therapeutic efficacy.2
The Ph is detected by G-band karyotyping in around 90% of CML patients among whom 5–10% may have variant types.3–5
Variant Ph chromosomes are characterized by the involvement of another chromosome in addition to chromosome 9 or 22. It can be a simple type of variant when only one additional chromosome is involved, or complex, in which two or more chromosomes, besides chromosomes 9 and 22, take part in the translocation.6,7
Variant Ph breakpoints occur in hotspots across the genome, usually in the G-light bands, within the cytosine and guanine (CG) rich parts of the genome.4 CG content correlates with chromatin condensation and transcription activity; that is, open chromatin is transcriptionally active and relatively likely to undergo breakage and repair with a consequent tendency to illegitimate recombination and translocation.4 The 3′ portion of the BCR gene recombines preferentially with Alu elements at the breaks.8 However, the mechanism of variant Ph generation and the molecular bases of biological differences between classic Ph and variant Ph chromosomes are not fully understood.9 Recently, Albano et al.10 reported a study they performed on gene expression profiling (GEP) using microarrays to identify some of these differences. They found 59 genes showing differential expressions. Some of them (TR1B1, PTK2B and C5AR1) were involved in the MAPK pathway, which is already known to play a role in CML pathogenesis. The prognostic significance of variant Ph chromosomes has already been discussed,11 and it has been shown that the variant aberration does not impact on cytogenetic or molecular responses or even on clinical outcome.5
Variant Ph chromosomes are distinguished from additional chromosomal abnormalities or clonal evolution that drives disease progression. The clonal evolution is a reflection of a genetic instability that characterizes the transition to advanced phase.12 In this situation, i(17q), a second Ph and +8 are frequently found.
Few studies have reported the incidence of variant Ph chromosomes, or the breakpoints involved, in Brazilian CML patients.13,14
ObjectiveThe aim of this report is to describe the diversity of variant Ph chromosomes found in a group of Brazilian CML patients and to highlight some interesting breakpoint candidates for further studies.
MethodsA search was conducted of variant Ph chromosomes among all the cases of CML in the Cytogenetics Section Database of Fleury Laboratory (São Paulo, Brazil), during a 12-year period (from 1999 to 2010). The diagnosis of CML was made according to WHO Tumor Classification,1 with analysis of complete blood count, marrow aspiration and/or biopsy and karyotyping. Acute myeloid or lymphoid leukemia cases were excluded.
Karyotype analyses were performed from marrow samples, through non-stimulated short term cultures and classified according to the International System for Human Cytogenetics Nomenclature (ISCN).15 CML cases with positive BCR-ABL1 detection by fluorescent in situ hybridization (FISH) or polymerase chain reaction (PCR) at diagnosis, but with cryptic rearrangements by karyotyping, were excluded from the study since they were cytogenetically invisible.
The study was approved by the Institution Ethics Committee.
ResultsFifty cases (5.17%) of variant t(9;22) were found out of 1071 Ph-positive CML. Table 1 shows the list of the variant Philadelphia translocation cases (some of these cases were previously reported16) and Table 2 shows the rearrangement breakpoints. A total of 33 different regions and almost all chromosomes were involved in rearrangements, except for chromosomes: 8, 9, 18, 21, 22, X and Y. In two cases it was not possible to identify the extra chromosome of the variant translocation.
List of the variant Philadelphia translocations found.
| Case | Karyotype | Type of translocation |
|---|---|---|
| 1 | 46,XY,t(1;9;22;16)(q32;q34.1;q11.2;p13)[2]/46,XY | Complex |
| 2 | 46,XX,t(9;22;1;13)(q34.1;q11.2;p12;q34)[17]/46,XX[3] | Complex |
| 3 | 46,XY,t(9;22;1;?3)(q34.1;q11.2;q36.3;?q26)[20] | Complex |
| 4 | 46,XX,t(1;9;22;14)(p21;q34.1;q11.2;q32)[20] | Complex |
| 5 | 46,XX,t(9;22;1)(q34.1;q11.2;p34)[20] | Simple |
| 6 | 46,XX,t(9;22;1)(q34.1;q11.2;p36.3)[19]/46,XX[1] | Simple |
| 7 | 46,XX,t(9;22;2)(q34.1;q11.2;q37)[20] | Simple |
| 8 | 46,XY,t(2;9;22)(p21;q34.1;q11.2)[20] | Simple |
| 9 | 46,XY,t(9;22;2)(q34.1;q11.2;q24)[8]/46,XY[12] | Simple |
| 10 | 46,XY,t(9;22;4)(q34.1;q11.2;p16)[20] | Simple |
| 11 | 46,XY,t?(9;22;10;11)(q34.1;q11.2;q22;q25)[20] | Complex |
| 12 | 46,XY,t(9;22;5)(q34.1;q11.2;p13)[20] | Simple |
| 13 | 46,XY,t(9;22;6)(q34.1;q11.2;q22) | Simple |
| 14 | 46,XX,t(9;22;6)(q34.1;q11.2;q27)[7]/46,idem,?del(12)(p11.2p13)[3]/46,XX[10] | Simple |
| 15 | 46,XY,t(9;22;6)(q34.1;q11.2;q22) | Simple |
| 16 | 46,XY,t(9;22;15)(q34.1;q11.2;q11.2)[20] | Simple |
| 17 | 46,XX,t(9;22;7)(q34.1;q11.2;p22)[20] | Simple |
| 18 | 46,XX,t(?9;22;7)(q34.1;q11.2;q36), 9phqh[14]/46,XX, 9phqh[6] | Simple |
| 19 | 46,XY,t(9;22;10)(q34.1;q11.2;q21)[17]/46,XY[3] | Simple |
| 20 | 46,XY,t?(9;22;10;11)(q34.1;q11.2;q22;q25)[20] | Complex |
| 21 | 46,XY,t(9;22;4)(q34.1;q11.2;p16)[1]/45,idem,add(7)(p22),-8[18]/46,XY[1] | Simple |
| 22 | 44∼45,XX,t(7;12)(q11.2;p12),?t(9;22;11)(q34.1;q11.2;p15),-14,-15,1min[cp18]/46,XX[2] | Simple |
| 23 | 46,XY,t(9;22;11)(q34.1;q11.2;p15),9ph[20] | Simple |
| 24 | 46,XX,t(9;?22;12)(q34.1;?q11.2;q24)[17]/46,XX[3] | Simple |
| 25 | 46,XY,t(1;7)(p22;q22),t(9;22;20)(q34.1;q11;q11.2)[4]/46,XY,t(9;22;20)(q34.1;q11;q11.2)[3] | Simple |
| 26 | 46,XY,t(9;22;12)(q34.1;q11.2;p13)[20] | Simple |
| 27 | 46,XY,t(9;22;14)(q34.1;q11.2;q32)[19]/46,XY[1] | Simple |
| 28 | 46,XY,t(9;22;15)(q34.1;q11.2;q25),t(17;19)(q12;q13.4)[20] | Simple |
| 29 | 46,XY,t(9;22;15)(q34.1;q11.2;q11.2)[20] | Simple |
| 30 | 46,XX,t(9;22;6)(q34.1;q11.2;q27)[7]/46,idem,?del(12)(p11.2p13)[3]/46,XX[14] | Simple |
| 31 | 46,XX,t?(9;22;16)(q34.1;q11.2;p13.3)[20] | Simple |
| 32 | 46,XX,t(9;22;17)(q34.1;q11.2;q21?)[14]/46,XX[6] | Simple |
| 33 | 46,XX,t(9;22;17)(q34.1;q11.2;q?21)[16]/46,XX[4] | Simple |
| 34 | 46,XY,t(9;22;17)(q34.1;q11.2;q21)[20] | Simple |
| 35 | 46,XX,t(9;22;17)(q34.1;q11.2),t(12;13)(q13;p11)[20] | Simple |
| 36 | 46,XY,t(9;22;17)(q34.1;q11.2;p13)[20] | Simple |
| 37 | 46,XX,t(9;22;17)(q34.1;q11.2;q24)[20] | Simple |
| 38 | 46,XY,t(9;22;17)(q34.1;q11.2;p13)[20] | Simple |
| 39 | 46,XX,t(9;22;17)(q34.1;q11.2;q21)[10]//46,XX[10] | Simple |
| 40 | 46,XY,t(9;22;17)(q34.1;q11.2;p13)[3]/75∼77,XY,+X,+1,+5,+6,+9,t(9;22;17)(q34.1;q11.2;p13)x2,+10,+11,+12,+20,+21,+22[cp16]/46,XY[1] | Simple |
| 41 | 46,XX,t(9;22;17)(q34.1;q11.2;q24)[1]/47,XX,+8[8]/46,XX[11] | Simple |
| 42 | 46,XY,t(1;7)(p22;q22),t(9;22;20)(q34.1;q11;q11.2)[20] | Simple |
| 43 | 46,XY,t(9;22;19)(q34.1;q11.2;q11.2)[20] | Simple |
| 44 | 46,XY,t(9;22;20)(q34.1;q11.2;q11.2)[13]/46,XY,t(1;7)(p22;q22),idem[7] | Simple |
| 45 | 46,XY,t(1;7)(p22;q22),t(9;22;20)(q34.1;q11;q11.2)[2]/46,XY[18] | Simple |
| 46 | 46,XX,t(9;22;17)(q34.1;q11.2;q24)[1]/47,XX,+8[11]/46,XX[8] | Simple |
| 47 | 46,XY,t(9;22;12)(q34.1;q11.2;q24.3)[20] | Simple |
| 48 | 46,XY,t(1;7)(p22;q22),t(9;22;20)(q34.1;q11;q11.2)[1]/46,XY[9] | Simple |
| 49 | 46,XY,t(9;22;?)(q34.1;q11.2;?)[11]/46,XY[9] | Simple |
| 50 | 46,XY,t(9;22;?)(q34.1;q11.2;?)[20] | Simple |
The incidence of variant Ph chromosomes in Brazilian CML patients was similar to published data (5.17%).17 Forty-four (88%) were simple variants and six (12%) complex (Table 1), also in accordance with previous reports.4
The most frequently involved chromosome was 17 (eleven times more common in the present report), followed by chromosome 1 (six times), 20 (five times), 6, 11 (four times each), and 2, 10, 12 and 15 (three times each). Marzocchi et al.5 reported four cases of the involvement of chromosome 17 out of 30 variants. The chromosomes not involved in Ph variant translocations in the present study were: 8, 9, 18, 21, 22, X and Y, while other studies have shown rearrangements of these chromosomes, except for Y.14,17,18
Among all breakpoints seen in this survey, six were repeated: 11p15, 14q32, 15q11.2, 16p13.1, 17p13 and 17q21, differing from the report by Marzocchi et al.5 who did not find any recurrent breakpoints. On the other hand, Reid et al.19 listed nine recurring breakpoints in variant Ph translocations, most of which had been reported by others. In fact, it seems that most breakpoints related to variant Ph chromosomes occur in regions of known oncogenes or typical secondary breakpoints in other types of cancer. Indeed, we endorse this statement, since the current study also identified some breakpoints common to other cancers, such as 12p13, 17p13 and 17q21.19 Some of the genes located in these regions are related to hematological malignancies (i.e. ETV6, CD9, RARA, GAS7, IGH-2, MYB). However, additional genes have already been reported in respect to CML, including TP53, located at 17p13, and involved in up to 30% of CML cases in blast crisis20 and MECOM and VRK1 that play a role in blast crises.21 Moreover, it is also known that the BCR-ABL1 tyrosine kinase activity activates the P13K/AKT signaling pathway.21 Thus, more studies regarding the function, expression and regulation of these and other genes may help to explain how variant Ph translocations occur.
Controversies were found in respect to the meaning of variant Ph chromosomes. Some authors have stated that the involvement of additional oncogenes could be associated to poorer prognosis although the European LeukemiaNet recommendations do not provide advice for patients with variant translocations.2,5 In the tyrosine kinase inhibitors era, it seems that variant Ph translocations have no implication on the prognosis, evolution of the disease and response to treatment.5,18
ConclusionsDue to the large variety of Ph variant translocations, it seems that many possible interactions exist with some occurring in regions already reported as hotspots of oncogenes or related to different types of cancer. This suggests that there must be local phenomena favoring the predisposition to the involvement of these breakpoints.
The incidence of Ph variant translocations in Brazilian patients is similar to other reports worldwide however some new variants were found. 17q21 was the most frequent and repeated breakpoint and so this region is the most likely to be involved in Ph variant translocations. Nevertheless, the pathological implication of these variants remains unclear.
Conflicts of interestThe authors declare no conflicts of interest.