One of the major challenges in genetic studies is to ascertain the pathogenicity of a given variant in order to assess its role in disease development and progression. In the era of genomic discovery, we have more questions than answers. A strategy to interpret the clinical significance of rare or novel variants appropriately is needed to avoid wrong clinical decisions based on common variants.
From the 1990s to 2010, the discovery of a genetic basis for Mendelian diseases was made primarily by conventional sequencing approaches, such as Sanger sequencing. In the past few years, the pace of genome screening increased with the introduction of the next-generation sequencing (NGS) technologies, such as whole-genome sequencing (WGS) or whole-exome sequencing (WES).1 The development of NGS has dramatically reduced the cost of sequencing and increased the amount of data, whereas the Sanger method was always costly and time-consuming. Combined, the WGS and WES have discovered more than the double of genes as conventional approaches.2 However, Sanger sequencing is still a powerful tool at different research centers in which NGS is not available or economically feasible. Regardless of the method employed, the discovery of novel variants that are etiologic in diseases is an essential piece of clinical research. The interpretation of the data obtained stands as the primary barrier to genetic screening rather than the mere discovery of novel variants.3
A recent review wisely stated that “damaging does not mean pathogenic”.3 Indeed, in silico approaches alone are not enough to assess the pathogenicity of novel variants and the “probably damaging” or “tolerated” results determined by SIFT and Polyphen software cannot reliably predict whether a variant is disease-causing. Different types of evidence have been used in the process of variant interpretation, and their use has been standardized by guidelines developed in Europe and the United States. The American College of Medical Genetics and the Association for Molecular Pathology (ACMG) issued a consensus guideline in 2015 that combined computational, functional, population and clinical data as criteria to stratify the strength of evidence and to determine the pathogenic status.4 This guideline standardized the interpretation of variants, workflows and improved the outcomes of the assessment of pathogenicity of novel variants reported across studies.
In this issue of the Brazilian Journal of Hematology and Hemotherapy, Svidnicki et al. identified variants in the PKLR gene associated with pyruvate kinase deficiency and used the ACMG criteria to evaluate their clinical significance.6 The study contributes to the molecular characterization of a recessive disorder in a population with a complex genetic background that has not been screened in the same proportion as the American or European populations. The South American population, specifically, the Brazilian population, is underrepresented in most of the genome databases. The likely pathogenic R486W PKLR variants identified in three unrelated patients by Svidnicki et al. have a mean allele frequency of 0.28% in the ExAC Exome Aggregation Consortium (ExAC – http://exac.broadinstitute.org) or 0.30% in the Genome Aggregation Database (GnomAD – http://gnomad.broadinstitute.org). However, the Latino population represents only 27% of the individuals screened for this variant. Svidnicki et al. identified the R486W PKLR variant in 0.1% of their controls.6 In the ABraOM consortium database (http://abraom.ib.usp.br/) the same variant has a frequency of 0.32%. The ABraOM is a pioneering initiative attempting to overcome this limitation by providing genetic variability among Brazilians.5 Indeed, the frequency of the R486W PKLR variant is relatively high in all the databases evaluated, however, this is common for a recessive disease-causing variant.
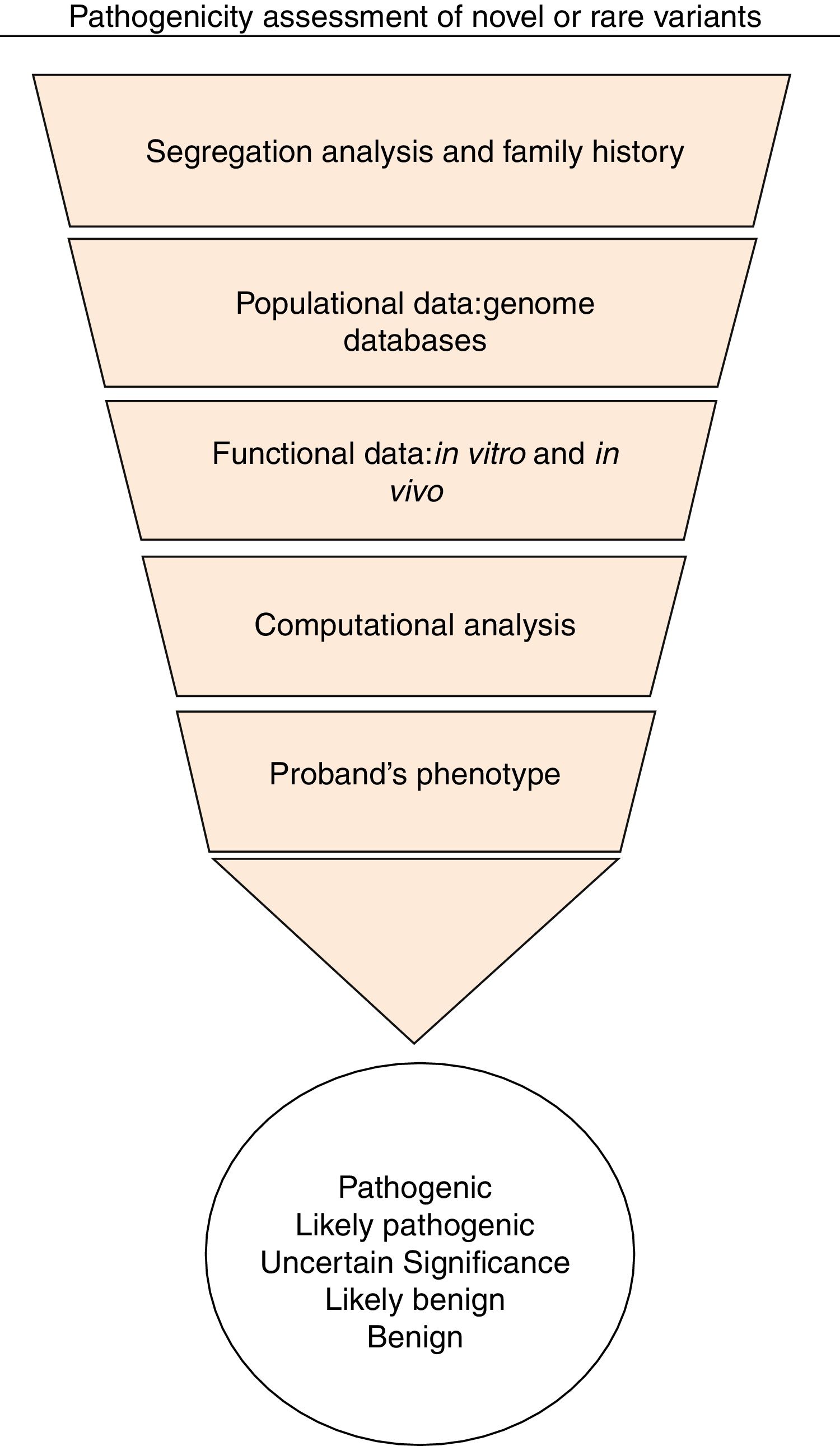
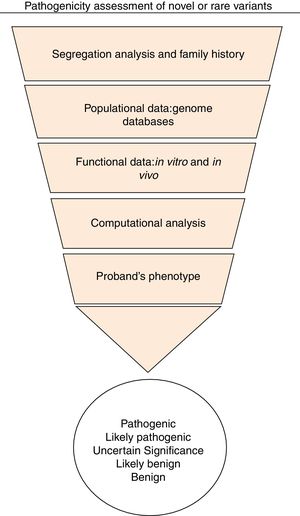
From a practical standpoint, the NGS approach identifies many rare variants whose pathogenicity will remain unclear in the absence of further analysis. Following the ACMG criteria, Svidnicki et al., 2017 classified three novel variants as of uncertain significance, probably due to the lack of functional or family history data, the main limitations in the process of assessing the pathogenicity of novel variants.6 In general, the combination of different tools for in silico prediction, well-established criteria to classify variants, a variant allele frequency control from ExAC or GnomAD, familial investigation, and functional data are critical to increase the reliability of sequencing results and to provide evidence for pathogenicity (Figure 1).
The ACMG criteria combined the following evidence to classify variants: population frequencies of variants in genome databases, computational and in silico predictions, functional data, segregation analysis from family pedigrees, allelic data, functional data, and patient's phenotype. Variants are classified as pathogenic, likely pathogenic, of uncertain significance, likely benign, or benign.
Conflicts of interestThe authors declare no conflicts of interest.
See paper by Svidnicki et al. on pages [5–11].