T315I mutations of the BCR::ABL1 gene lead to resistance to tyrosine kinase inhibitors (TKIs). This study evaluated the performance of digital droplet polymerase chain reaction (ddPCR) in quantifying T315I mutations and their frequency in Philadelphia chromosome (Ph) positive hematological patients.
MethodsThe course of disease and BCR::ABL1 fusion transcripts (e13a2, e14a2 and e1a2) were retrospectively reviewed in 21 patients with acute lymphoblastic leukemia (ALL) and 85 patients with chronic myeloid leukemia (CML). T315I mutation analysis was carried out using ddPCR and the limit of detection was assessed using mutant T315I DNA at varying variant allele fractions.
ResultsT315I mutations were found in two ALL patients and one CML patient without remission in molecular biology and with mutation burdens of 29.20%, 40.85%, and 3.00%, respectively. The mutation burden of ALL patients was higher than that of CML patients, but there was no significant difference between the two (p-value = 0.0536). The test's limit of detection was 0.02% with a correlation coefficient greater than 0.99 between the expected and actual detection abundances.
ConclusionT315I mutations have a high incidence in Ph-positive ALL patients even if the course of disease is short. In molecular biology, T315I mutation detection is indicated for CML patients not in remission.
The Philadelphia chromosome (Ph) is a result of the reciprocal translocation between chromosome 9 and 22, t(9; 22)(q34; q11), resulting in the formation of the BCR::ABL1 fusion gene that encodes tyrosine kinase (TK). This naturally active TK activates multiple signal transduction pathways leading to uncontrolled proliferation. In more than 95% of chronic myeloid leukemia (CML) cases, the BCR::ABL1 transcript results from the fusion of exon 13 or 14 of the BCR gene and exon 2 of the ABL1 gene (e13a2 and e14a2, respectively), yielding a 210 kDa protein product.1 In Ph-positive acute lymphoblastic leukemia (ALL), the fusion of exon 1 of the BCR gene and exon 2 of the ABL1 gene (e1a2) is the major isoform and encodes a 190 kDa protein product.2
The advent of BCR::ABL1 tyrosine kinase inhibitor (TKI) has revolutionized the therapeutic prospects of CML. However, drug-resistant mutations in BCR::ABL1 are inevitable, which presents considerable challenges for current targeted therapies. More than 90 types of mutations in the BCR::ABL1 kinase region have been identified, including T315I, E255K, Y253F, Q252R, G250E, L248V and M244V.3 T315I is one of the most common drug-resistant mutations in BCR::ABL1 accounting for approximately 20% of clinical resistance to TKIs.4 The occurrence of T315I mutations in BCR::ABL1 results in resistance to most of the TKIs currently used, including first-generation imatinib and second-generation nilotinib.5 Mutation testing is recommended in case of poor or failed treatment response to optimize the selection of the different TKIs.6
Previous studies have analyzed T315I mutations through direct sequencing,7 restriction fragment length polymorphism8 and or fluorescence in situ hybridization.9 However, these methods are time-consuming and laborious. Although fluorescence PCR detection of T315I mutations is a convenient and sensitive method, it is mostly used for qualitative detection.6 The latest ‘version’ of quantitative PCR is ddPCR technology which offers nanoscale PCR with water in oil droplets generated during the partitioning process and performs Poisson statistics to estimate the absolute amount of target DNA in the original sample.10 A calibration curve is not required for absolute quantification of unknown samples. In addition, ddPCR is more sensitive and accurate than quantitative PCR.11 In view of these advantages, many biotechnology companies have produced different digital PCR platforms, such as QX 200, Giant Studio 3D, QiAcuity and SG-2000. Currently, ddPCR is being applied to various tests, such as detection of pathogens in patients with bloodstream infections12 and mutation detection in circulating tumor DNA.13
This study aimed to evaluate the frequency of the BCR::ABL1 T315I mutations in patients with Ph-positive ALL and in different categories of CML. This study also aimed to evaluate the sensitivity and accuracy of this method for screening BCR::ABL1 mutations on the SG-2000 digital PCR system.
Materials and methodsStudy populationThis retrospective study was conducted at the Affiliated Wuxi People's Hospital of Nanjing Medical University. A total of 106 Ph-positive patients were included in the present study. Based on the results of BCR::ABL fusion gene testing, 33 patients with CML were in molecular remission (MR) and 52 patients with CML were not in MR. For 21 patients with ALL, 17 were not in MR and four in MR. All CML patients in the study cohort had a disease course of over one year, whereas ALL patients were included regardless of their disease course. Among them, 44 patients received treatment with the first-generation TKI imatinib, while 34 patients received treatment with second-generation TKIs, including nilotinib, dasatinib and flumatinib. The remaining 28 patients were treated with hydroxyurea or their medication status was unclear. The Ethics Committee of Wuxi People's Hospital of Nanjing Medical University approved this study (Permit number: KY23044) and all protocols followed the ethical guidelines of the 1975 Helsinki Declaration.
Genomic DNA extractionTo extract genomic DNA, the remaining white-colored interphase and the lower phenol-chloroform phase were used after RNA extraction by the TRIzol method using the magnetic bead technique. Briefly, 200 µL of a mixture of the interphase and the lower phenol-chloroform phase was pipetted into the first or seventh column of a pre-dispensed RNA/DNA 96 well extraction plate (Tianlong Technology Co., Ltd, Xi'an, China). The RNA/DNA extraction plates were placed in an automated nucleic acid extraction instrument (GeneRotex96, Tianlong Technology Co., Ltd, Xi'an, China). The 2D code of the RNA/DNA extraction kit was scanned to set up the extraction procedure. The purified DNA was located in columns 6 and 12. The concentration and purity of DNA were determined using a spectrophotometer (NanoDrop, USA) at 260 and 280 nm.
Quantitative measurement of T315I mutations by digital droplet PCRTo quantify T315I mutations using ddPCR, the samples were subjected to processing using the ddPCR BCR::ABL T315I kit (Rainsure Scientific Co., Ltd, China) following the manufacturer's instructions. The PCR reactions were performed in a 20-μL reaction system containing 10 μL ddPCR Master Mix, 1 μL each of T315I wild and mutant probes and an 8 μL DNA sample. The entire reaction mixture and 75 μL of droplet generation oil were loaded into a ddPCR plate. The plates were then placed in a Sample Prep Station (SG-2000, Rainsure Scientific Co. Ltd, China). After droplet generation, ddPCR was performed with the following program: pre-denaturation at 95°C for 10 min, 40 cycles at 94°C for 30 s and at 55°C for 60 s, followed by 98°C for 10 min and held at 20°C until further processing. After PCR, the plates were transferred to a Cartridge Scanner (Dscanner4-1000, Rainsure Scientific Co., Ltd, China) and absolute quantifications of the mutant (FAM channel) and wild-type (HEX channel) alleles were estimated using Gene Count Analysis V1.63. Based on the examination of the aggregated two-dimensional (2D) droplet plots, the threshold was manually set in regions with the lowest rainfall between clusters. Any droplets exceeding the threshold are considered positive, while droplets below the threshold are considered negative.
Limit of detection and quantitative linearity of T315I mutations by digital droplet PCRThe limit of detection (LOD) and quantitative linearity of the ddPCR assay were evaluated by running a mutation dilution series. The mutation dilution series was obtained by mixing mutated and wild DNA in order to mimic different mutation burdens, from 10%, 5%, 1%, 0.5%, 0.1% down to 0.02%. Each mutation burden sample was tested in triplicate.
StatisticsT-tests were used to analyze the differences in disease course between the MR and non-MR patients. The Mann Whitney U test was also used to examine the variation in mutation burden between the ALL and CML patients without remission in molecular biology. Correlation analysis between expected mutation abundance and detected mutation abundance was performed using the Excel data analysis function. Fisher's exact test was employed to analyze the difference in T315I mutation incidences between the ALL and CML patients.
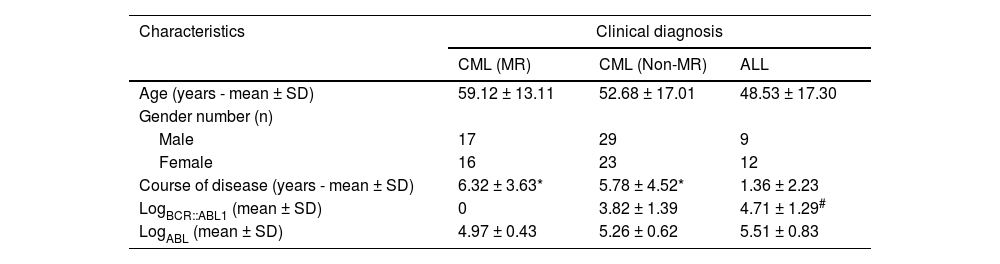
ResultsPatient characteristicsThe course of the disease and expression of the BCR::ABL1 fusion gene of the subjects in this study are shown in Table 1. All patients with CML had a disease course of more than one year with a maximum of 16 years. For 21 patients with ALL, 14 had a course of less than 12 months. There was a significant difference in the course of disease between the CML group and the ALL group (p-value <0.01). However, there was no significant difference in the disease course between the MR and non-MR CML patients. Among ALL patients, the BCR::ABL1 fusion genes encoding p210 and p190 accounted for 14.29% (3/21) and 85.71%, respectively. In CML patients, the BCR::ABL1 fusion genes encoding p210 and p190 accounted for 98.82% (84/85) and 1.18%, respectively.
Demographic and clinical characteristics.
| Characteristics | Clinical diagnosis | ||
|---|---|---|---|
| CML (MR) | CML (Non-MR) | ALL | |
| Age (years - mean ± SD) | 59.12 ± 13.11 | 52.68 ± 17.01 | 48.53 ± 17.30 |
| Gender number (n) | |||
| Male | 17 | 29 | 9 |
| Female | 16 | 23 | 12 |
| Course of disease (years - mean ± SD) | 6.32 ± 3.63* | 5.78 ± 4.52* | 1.36 ± 2.23 |
| LogBCR::ABL1 (mean ± SD) | 0 | 3.82 ± 1.39 | 4.71 ± 1.29# |
| LogABL (mean ± SD) | 4.97 ± 0.43 | 5.26 ± 0.62 | 5.51 ± 0.83 |
CML: chronic myeloid leukemia; MR: molecular remission; ALL: acute lymphoblastic leukemia.
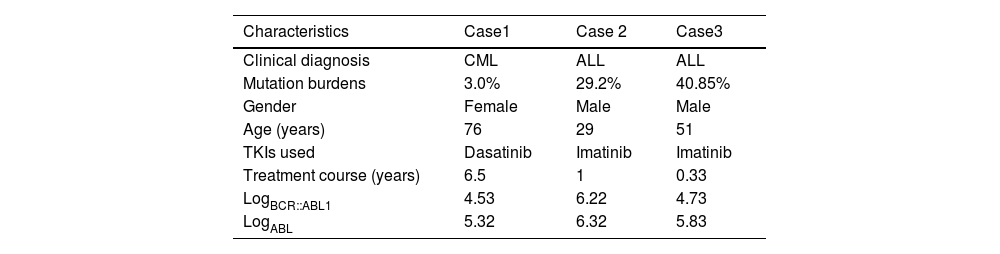
One mutation was detected in the non-MR CML group with a fractional abundance of 3.0% and two non-MR patients with ALL had T315I mutations with fractional abundances of 29.20% and 40.85%, respectively. No T315I mutation occurred in the MR patients with CML or ALL. The mutation burden of ALL patients was higher than that of CML patients, but there was no significant difference between the two (p-value = 0.0536). The frequencies of T315I mutations were 1.92% and 11.76% in non-MR patients with CML and ALL, respectively. There was no significant difference in the incidence of T315I mutations between ALL and CML patients (p-value = 0.13). Clinical characteristics of the three patients with T315I mutations are shown in Table 2.
Clinical characteristics of three patients with T315I mutations.
CML: chronic myeloid leukemia; ALL: acute lymphoblastic leukemia.
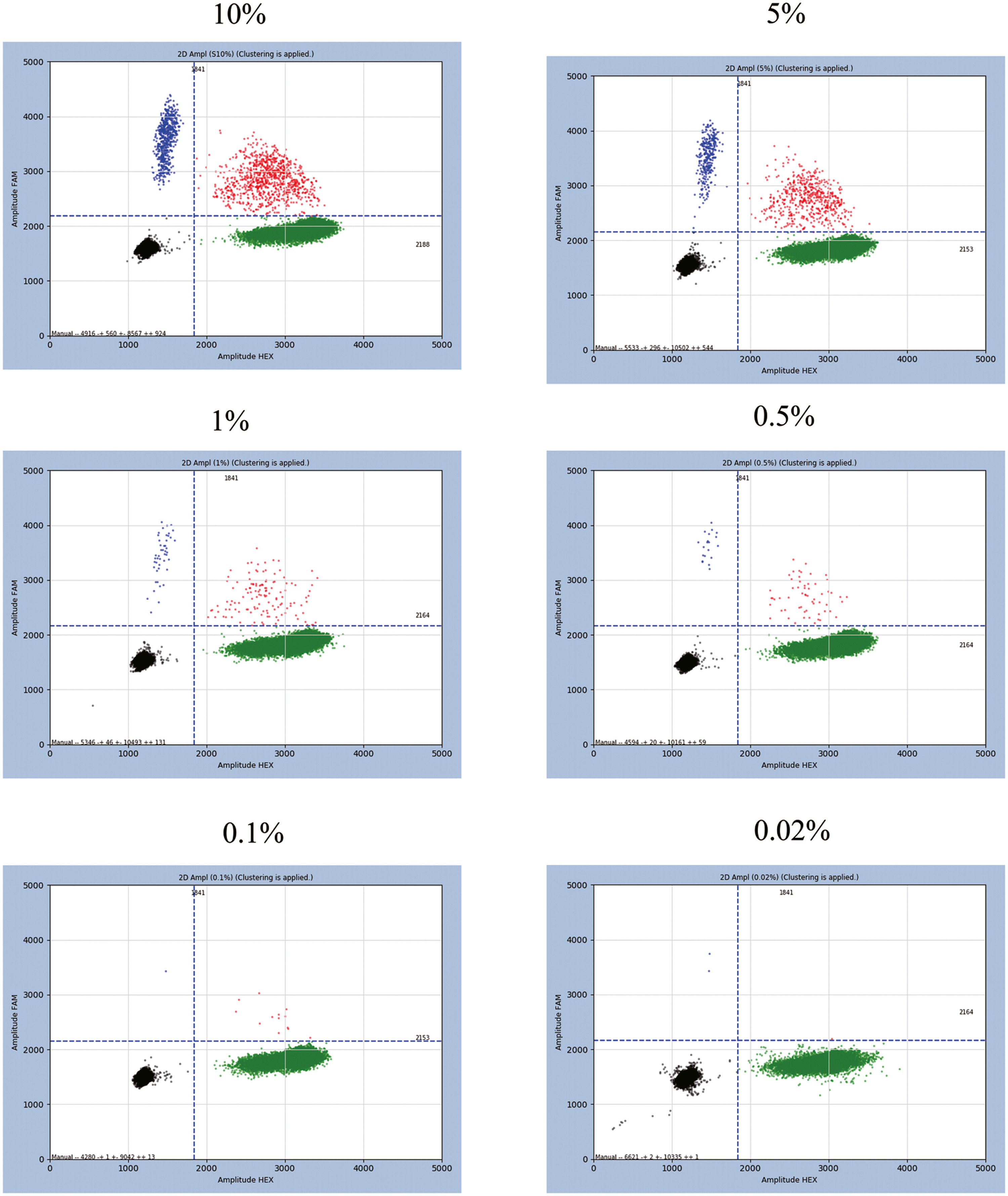
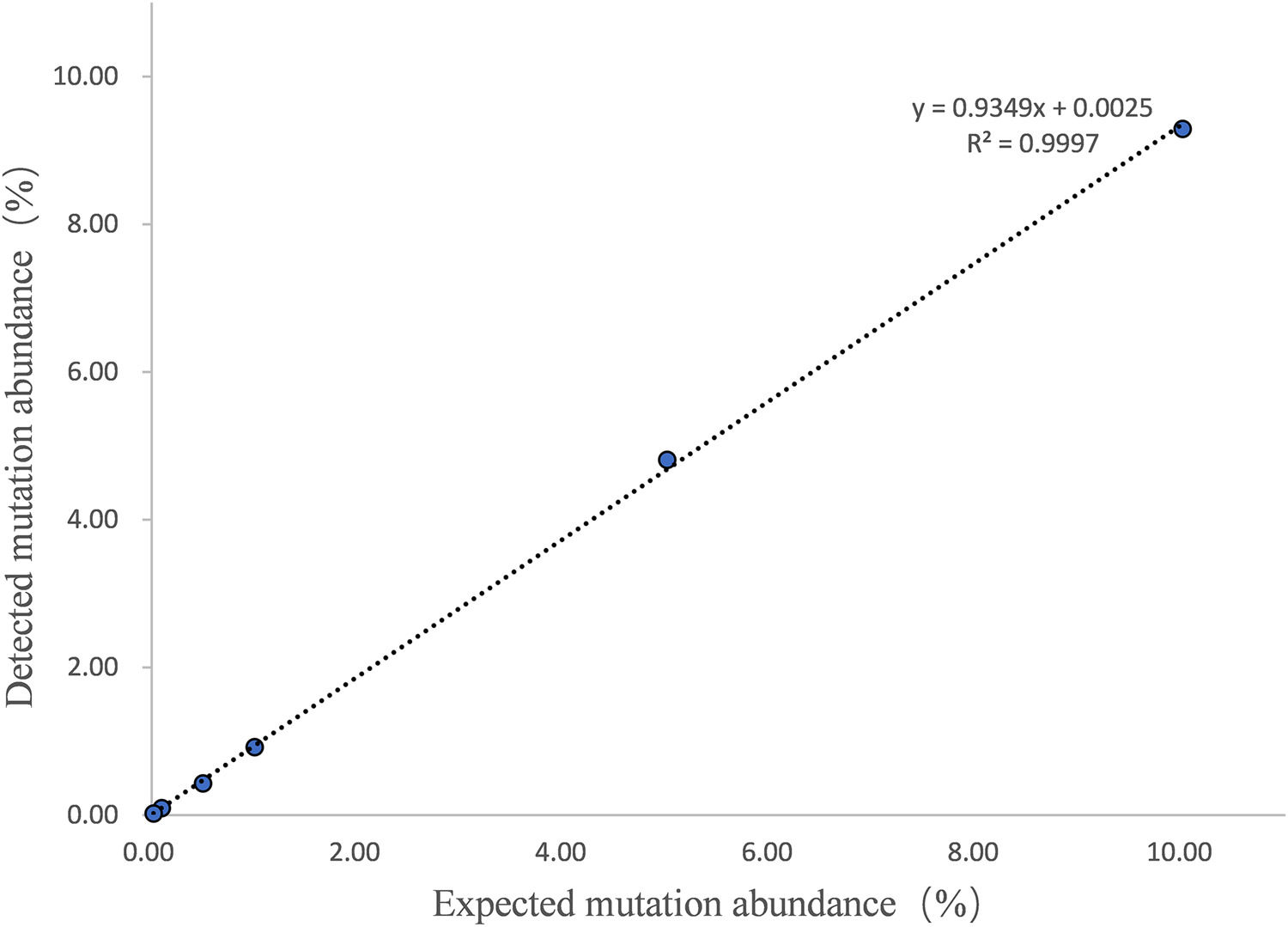
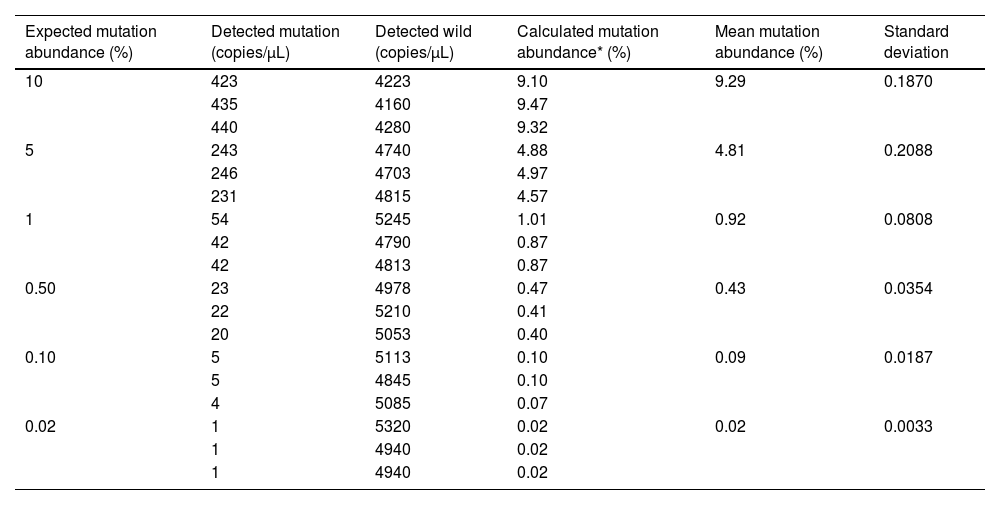
The LOD and quantitative linearity were investigated by running mutation titration series. A mutation titration series was obtained by mixing mutated and wild DNA samples as shown in Table 3. Figure 1 shows the 2D plots of the mutation series obtained by multiplex ddPCR for T315I mutations. Mutations were identified down to 0.02% abundance at 120 ng DNA input. Table 4 shows the calculated mutation abundance for the T315I mutation dilution series using this ddPCR assay. The data show good linearity between the expected and measured mutant frequencies with an estimated slope coefficient of 0.935 and R2 = 0.9997 (Figure 2).
Preparation of T315I series in DNA mutation abundance testing.
Two-dimensional (2D) plots obtained by the multiplex ddPCR for the T315I mutation at different percentages. Black clusters at the bottom left corner represent FAM/HEX double-negative droplets. Green clusters at the bottom right corner correspond to droplets positive for the BCR::ABL1 gene (detected by HEX-conjugated probes). Blue clusters at the top left corner correspond to droplets positive for the BCR::ABL1 fusion gene with the T315I mutation (detected by FAM-conjugated probes). Orange clusters at the top right corner correspond to double-positive droplets.
Number of positive mutant events in the T315I mutation dilution series.
| Expected mutation abundance (%) | Detected mutation (copies/µL) | Detected wild (copies/µL) | Calculated mutation abundance* (%) | Mean mutation abundance (%) | Standard deviation |
|---|---|---|---|---|---|
| 10 | 423 | 4223 | 9.10 | 9.29 | 0.1870 |
| 435 | 4160 | 9.47 | |||
| 440 | 4280 | 9.32 | |||
| 5 | 243 | 4740 | 4.88 | 4.81 | 0.2088 |
| 246 | 4703 | 4.97 | |||
| 231 | 4815 | 4.57 | |||
| 1 | 54 | 5245 | 1.01 | 0.92 | 0.0808 |
| 42 | 4790 | 0.87 | |||
| 42 | 4813 | 0.87 | |||
| 0.50 | 23 | 4978 | 0.47 | 0.43 | 0.0354 |
| 22 | 5210 | 0.41 | |||
| 20 | 5053 | 0.40 | |||
| 0.10 | 5 | 5113 | 0.10 | 0.09 | 0.0187 |
| 5 | 4845 | 0.10 | |||
| 4 | 5085 | 0.07 | |||
| 0.02 | 1 | 5320 | 0.02 | 0.02 | 0.0033 |
| 1 | 4940 | 0.02 | |||
| 1 | 4940 | 0.02 |
This study evaluated the use of ddPCR for the detection of T315I mutations and found that it has potential clinical utility in monitoring T315I mutations in patients with ALL and CML. The results show that T315I mutations had a high incidence in Ph-positive ALL patients even if the course of disease was short. In addition, patients with CML who were not in MR were indicators of T315I mutation detection. The frequency of T315I mutations varies depending on the type of leukemia and treatment history of patients. In patients with CML, the frequency of T315I mutations has been reported to be between 2 and 20%.14 In the present study, the frequency in patients with CML was 1.18% (1/85), which was lower than that reported previously. T315I mutations have also been detected in patients with newly diagnosed Ph-positive ALL with frequencies ranging from 3.13 to 6.25%.15,16 In this study, the frequency in ALL with a course of the disease within 12 months was 13.33% (2/15). The data of the current study show that T315I mutations occur more frequently in Ph-positive ALL than CML.
The use of ddPCR in this study has several advantages over other detection methods. The sensitivity and accuracy of ddPCR are higher than those of quantitative PCR, and it does not require a calibration curve for the absolute quantification of unknown samples.17,18 These characteristics make it an efficient and accurate method for detecting BCR::ABL1 mutations. The current study shows that the ddPCR detection of T315I mutations has high sensitivity with a LOD of 0.02%. This provides a prerequisite for early change in the treatment strategies to an effective TKI. The data also show that ddPCR detection of T315I mutations is highly accurate and can quantify the burden of T315I. These results demonstrate that the T315I mutation burden in ALL patients was higher than in the CML patient (29.20% and 40.85% vs. 3.0%), but there is no significant difference. These findings will contribute to the precise medical treatment of patients with leukemia.
This study is important because T315I mutations in BCR::ABL1 confer resistance to most currently used TKIs, such as first-generation imatinib and second-generation nilotinib.19 Mutation testing is recommended in case of poor or failed treatment response to optimize the selection of TKI.20 Therefore, the use of ddPCR to detect T315I mutations can help clinicians identify patients who are likely to be resistant to TKIs and thus require alternative treatments.
However, this study has some limitations. The sample size was relatively small and the study was conducted at a single center. Therefore, the results may not be generalizable to other populations. In addition, the study did not investigate other mutations of the BCR::ABL1 gene, which can also cause drug resistance. Future studies should investigate the use of ddPCR to detect other mutations in BCR::ABL1.
ConclusionsIn summary, ddPCR is an efficient and accurate method to detect T315I mutations in the BCR::ABL1 gene in Ph-positive hematological patients. This study provides valuable information for clinicians to identify patients who are likely to be resistant to TKIs and require alternative treatments. Future studies should investigate the use of ddPCR to detect other mutations in the BCR::ABL1 gene to optimize treatment selection.