Recent advances in chronic lymphocytic leukemia (CLL) includes description of disease genomic landscape, inclusion of prognostic relevant genetic tests in CLL workflow and evaluation of minimal residual disease (MRD)1 in parallel with the increase availability of novel therapy agents.
In this review, the theoretical and practical aspects of response assessment have been discussed. These are based on updated recommendations of the European Research Initiative on Chronic Lymphocytic Leukemia (ERIC) for genetic tests (TP53 mutation and IGHV status) and flow cytometry analysis for CLL. Methodological approaches and interpretation of results were also discussed.2,3
CLL shows a heterogeneous clinical course encompassing a wide spectrum of different clinical presentations because some patients require early treatment while others exhibit an indolent course that does not affect their life span. Clinical heterogeneity is a reflex of differences in disease biology.1
CLL is a mature B-cell lymphoid neoplasm. Each normal mature B cell has a unique antigen-binding site as a result of random somatic rearrangement, a process occurring throughout B cell differentiation. In order to assemble a mature gene, somatic recombination of V(D)J segments of the heavy (IGH) and light (IGL) chain are rearranged to form the variable portion of the B cell receptor (BCR) in the mature B cell. Until now, no specific genetic abnormality has been associated to CLL, although BCR plays a crucial role in the survival of CLL cells.2
Chronic lymphocytic leukemia: diagnosis and differential diagnosis with other B-LPDDiagnostic criteria for diagnosing CLL rely on morphology and immunophenotyping. B-cell monoclonal lymphocytosis (MBL) may virtually precede all cases of CLL and small cell lymphocytic lymphoma (SLL). MBL is defined whenever a monoclonal B-cell population in peripheral blood (PB), likely or unlikely associated with a B CLL phenotype, is detected in conjunction with an absolute lymphocyte count <5000/mm3. Diagnostic criteria for CLL require an absolute lymphocyte count (ALC) ≥5000/mm3. Clonality may be identified either by flow cytometry or molecular assays. Flow cytometry analysis of mature B cell neoplasms identifying surface protein markers is generally the first and often a sufficient step for a diagnostic workup.3,4
A consensus criterion, based on a set of markers, is recommended for CLL diagnosis. Expression antigen profiles, assayed with required markers in peripheral blood samples, include at least CD19, CD5, CD20, CD23, Kappa, and Lambda. CLL is characterized by a clonal expansion of B cells with characteristic co-expression of CD19, CD5, CD23 and weak CD20, as well as sIg (surface immunoglobulin).
“Atypical” CLL is denoted when a CD19+ CD5+ phenotype shows ≥1 additional atypical CLL marker, like CD23+ and CD20 with weak sIg, CD23+ and CD20 with moderate sIg, or CD23−. A CCND1-IGH translocation occurs in 15%, 12% and 38% in these three atypical CLL phenotypes, respectively, while an MYD88 L265P mutation is detected in approximately 22–28% of these phenotypes.5,6
Differential diagnosis of CD5+ B− lymphoproliferative disorders (B-LPD) was often based on an immunophenotypic scoring system (Matutes score) comprising 5 markers: sIg, CD5, CD23, FMC7 and CD22. This score varied from 0 to 5 and was calculated assigning 1 point for expression of CD5 or CD23, 1 point for weak or lack of expression of either CD79b, sIg or CD20.7 However, differential diagnosis of CD5+ B− lymphoproliferative disorders (B-LPD) can be difficult due to significant overlap in clinical, cellular/immunophenotypic and molecular features within B-LPD. European Research Initiative on CLL (ERIC) recommendations for flow cytometry include additional markers like CD43, CD79b, CD81, CD200, CD10 and ROR1 for differential diagnosis and subsequent detection of minimal residual disease (MRD). However, the use of CD22, CD38, CD45 and FMC7 as complementary markers was not an unanimous choice.5
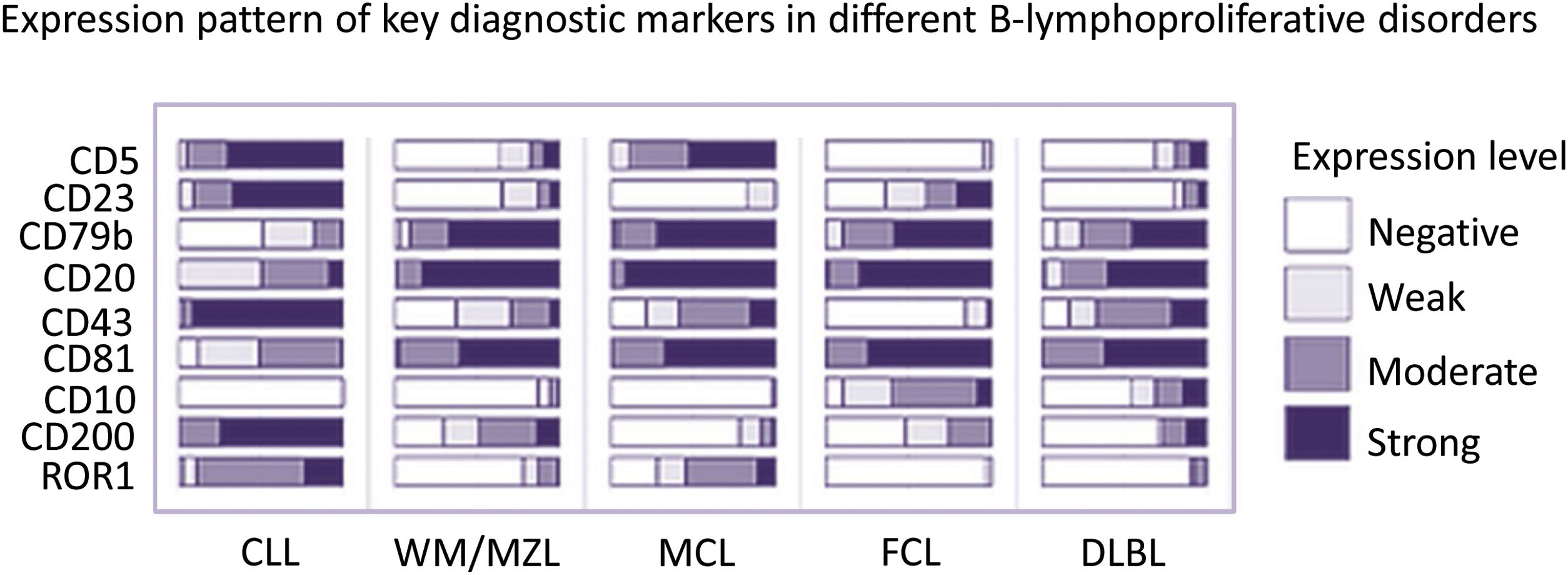
Based on the ERIC panel, another immunophenotypic scoring system was later proposed. This score, including 9 markers (CD5, CD23, CD79b, CD20, CD43, CD81, CD10, CD200 and ROR1) allowed for a differential diagnosis of different CD5 + B-LPD. The following CD5+ B-LPD categories can be discriminated: CLL, Waldestrom (WM) - Marginal Zone lymphoma (MZL), Hairy Cell Leukemia (HCL), Folicular Lymploma (FL) and Diffuse large B cell lymphoma (DLBL)5,6 (Figure 1).
Differential diagnosis of CD5+ B-lymphoproliferative disorders.
Expression pattern of Key diagnostic markers in different CD5+ B-lymphoproliferative disorders.
CLL, Chronic lymphocytic leukemia; WM, Waldenström’s Macroglobulinemia; MZL, Marginal Zone B-cell Non-Hodgkins Lymphoma; MCL, Mantle cell lymphoma; FCL, Follicle center lymphoma; DLBL, Diffuse Large B-Cell Lymphoma.
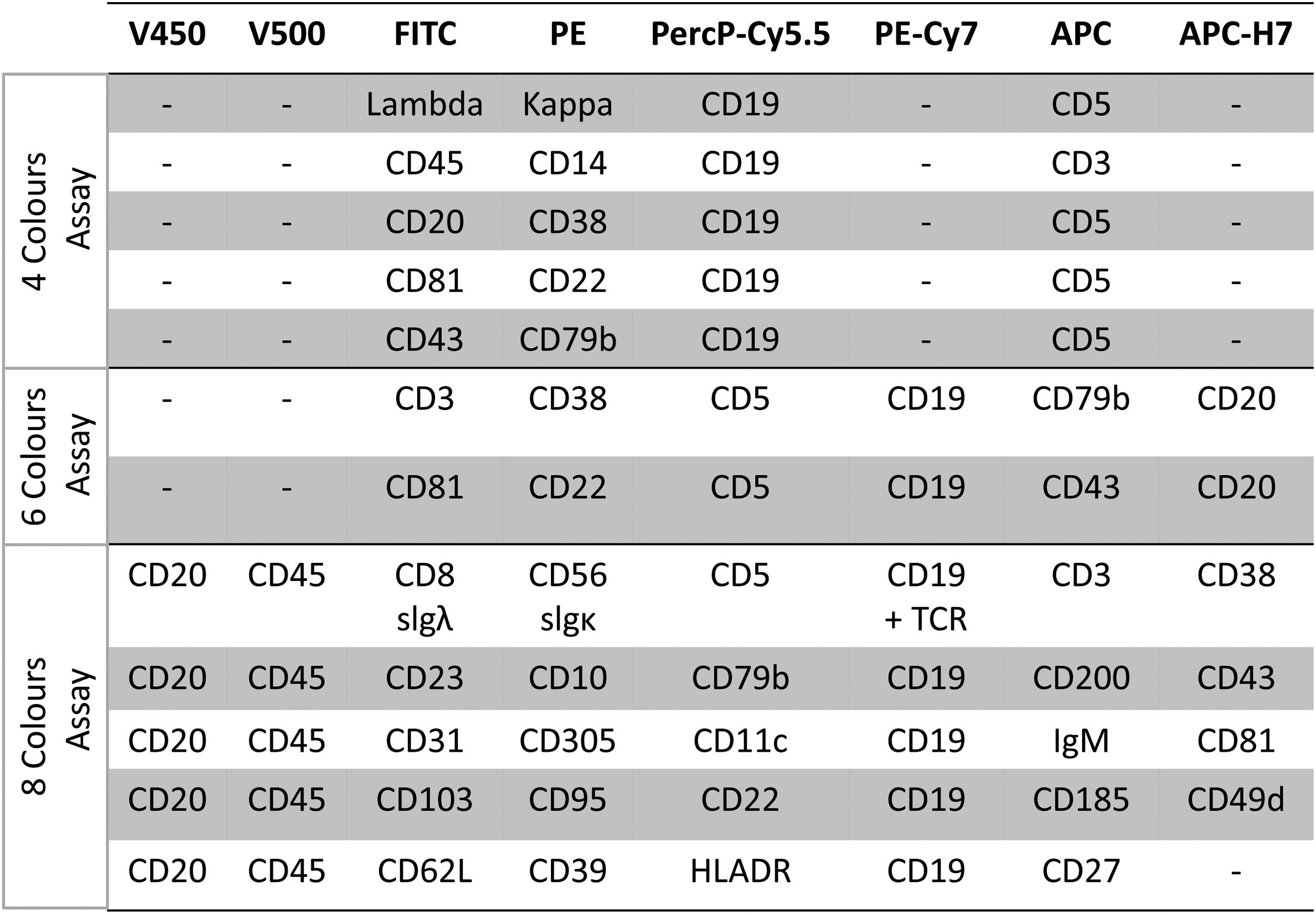
Multiparameter flow cytometry was recently validated by ERIC for quantifying residual cells (MRD) and as an extension of previous approaches. This assay is carried out in a single tube filled with a panel of six markers (CD19, CD20, CD5, CD43, CD79b and CD81) with a sensitivity below 0.01% (10–4). This process was initially standardized by reference laboratories and subsequently extended to other laboratories, including the testing of different platforms (FC500 cytometer- Beckman-Coulter) or FACSCanto II (BD Biosciences) and exchange of laboratory expertise. Harmonization was carried out focused on developing standardized kits containing 8 reagents (CD19, CD5, CD20, CD43, CD79b, CD81, CD200 and ROR1) in a fixed tube for prospective evaluation studies. Limits of detectability (LoD), quantificability (LoQ) and linearity were determined. Linearity (1.0) of findings between sample dilution tests, using a single tube with the 8-marker panel with limit of detection (LoD) of 10–5 or 0,001, showed a good concordance at the 0,01% of LoD. At present, the 0.001% (10–5) level is the threshold for classifying samples without detectable MRD. It was also demonstrated by ERIC that, for achieving a LoD of 0.001% (10–5) and a LoQ of 0.0025% (2.5 × 10–5), it was necessary to acquire 2 million events/tube, based on identification of CLL-phenotype cells above the 20 and 50 event thresholds respectively.8
In order to extend MRD harmonization to all laboratories and to standardize findings and criteria each laboratory should perform assays on its own to set limits of detection and, more importantly, to validate its LoQ to define its range of quantification for clearly scoring MRD levels, as well as non-quantifiable MRD (out of quantification range) and non- detectable MRD.8
In summary, the 2016 ERIC recommendations for MRD flow cytometry was an update of previous flow cytometry based on 4-color (in 4 tubes) and 6-color (in 2 tubes) panels. The later ERIC recommendations for detecting MRD is based on a core panel of six markers (CD19, CD20, CD5, CD43, CD79b and CD81) or 8 markers (CD19, CD5, CD20, CD43, CD79b, CD81, CD200 and ROR1) with component specifications regardless of platforms and reagent brands, which can be locally re-validated with normal peripheral blood. This approach shows linearity to a detection limit of 10−5. To achieve this level of sensitivity (0.001%), the acquisition of at least 500,000 cells is required. A sequential gating strategy for detecting a typical B-CLL phenotype and quantifying small residual B-CLL populations after treatment allows an effective distinction from normal polyclonal and regenerating B-cells (hematogones)8 (Figure 2).
Prognostic stratificationBased on the original study of Döhner, with fluorescence in situ hybridization (FISH) on interphase nuclei, risk-adapted treatment strategies based on genetic subgroups was proposed.9 Approximately 80% of CLL patients carry at least one of four recurrent chromosome gains or deletions (del). Most commonly, del(13)(q14.1) (40–50% of cases, RB1 and DLEU2 genes) followed by trisomy 12 (10–20% of cases), del(11)(q22-23) (15–20%, ATM gene) and del(17)(p13.1) (5–7%,TP53 gene).10
Risk stratification of CLL patient is based on these alterations. Patients with del17p show the worst prognosis followed by those with del11q, 12q trisomy, and with a normal karyotype. Patients with 13q deletions as the sole abnormality show the longest estimated survival. Testing these alterations with FISH probe panels containing D13S319/CEP12/13q34 and TP53/ATM at diagnosis/presentation are recommended by ERIC to all CLL patients. FISH analysis, carried out with a probe panel for detecting the common recurrent abnormalities, has become part of routine clinical evaluation at the time of CLL diagnosis.11
FISH analysis detects only specific alterations. In contrast, cytogenetic analysis can be successfully performed in metaphase nuclei activated with B cell mitogens, cytokines alone or in combined protocols. Lately, the importance of conventional cytogenetics in CLL workflow has been highlighted although is becoming clear that complex karyotypes result from TP53 loss of function.12–14
Microarray analysis has also been used to evaluate copy number variation in CLL patients. Interestingly, whole genome microarray testing reveals two types (I and II) of 13q14 deletions. Type I deletions are small, present in 30% of CLL cases associated to good prognosis, while Type II deletions are large and present in 20% of CLL cases associated to high genomic complexity and contributing to CLL evolution. The clinical outcome of CLL is accelerated in patients with type II 13q14 deletions spanning to the RB1 gene. Finally, complex array profiles as well as complex karyotypes have been associated to adverse prognosis. It is worth noticing that genomic complexity can also be defined by microarray.15,16
Recurrent mutations refines prognosis in chronic lymphocytic leukemiaOver the past 12 years, re-sequencing studies identified a number of biologic markers associated to CLL. Screening for NOTCH1, SF3B1, TP53, MYD88 and BIRC3 mutations at diagnosis and before treatment supported the clinical relevance of testing these recurrent mutations in CLL. The 2016 WHO classification for lymphoid neoplasms already ascribes TP53, NOTCH1, SF3B1, ATM and BIRC3 mutations to CLL. Moreover, the adverse impact of NOTCH1, SF3B1 and TP53 mutations, independently of the IGHV mutational status, is presently highlighted.17 Standardization/harmonization of NGS technology for clinical use will further contribute to the assessment of the prognostic significance of these alterations.18
The introduction of molecular genetics for risk stratification models refined CLL prognosis. Recurrent mutations in NOTCH1, SF3B1 and, mainly, the mutational status of the variable immunoglobulin heavy chain (IGHV) genes were the most important markers for prognosis and predicting the timing for initiating treatment.11
TP53 alterations (TP53alt: mutations and/or deletions) and unmutated IGHV (U-CLL) were identified as the most significantly adverse prognostic factors followed by SF3B1 mutations, especially when associated with del(11q), isolated del(13q) or expressing unmutated IGHV genes. Although recurrent mutations seem to be important determinants of prognosis, the incorporation of IGHV mutational status and TP53alt are still the only genetic markers, apart from Döhner´s FISH prognostic model, included in the 2018 iwCLL guideline.11
TP53 alterations (del17p or TP53 mutation)Loss of TP53 function may occur due to del(17p) or mutation. Most often, del(17p) is associated to a TP53 mutation on the remaining allele although TP53 mutations can occur in 5% of untreated patients without del(17p). Approximately 90% of patients with del(17p) carry a TP53 mutation; conversely, only 60–70% of patients with a TP53 mutation also harbor del(17p) detected by FISH.
Most TP53 mutations are missense and localized at the DNA-binding core domain. Despite that some codons are more frequently mutated (hotspots), deleterious variants may occur in any part of the coding sequence. In CLL, TP53 mutations are frequently subclonal and clinically significant, even if the other allele remains intact. TP53 mutations are detected with an increased sensitivity by using purified B cells from a mononuclear cell gradient. Information on blood cell count parameters (at least WBC) provided with the sample is helpful for handling laboratory samples.19,20
The 2018 ERIC recommendations for analyzing TP53 mutation in CLL updated previous methodological approaches and interpretation of findings. Briefly, they included recommendations for an appropriate initial handling of samples (mononuclear or purified B cells), regions of interest (at least exon 4–10 and, optimally, the complete coding region from exon 2–11 including intronic flanking regions) and protocols and quality parameters for Sanger or next-generation sequencing (NGS). The recommended threshold for reporting mutations detected by NGS should reflect the Sanger-like threshold of ∼10% of the variant allele. Examples of molecular findings and clinical reports are discussed in this latest ERIC recommendation.19
Predictive implications of TP53 inactivation have an increasing significance because new target therapies based on BTK inhibition and BCL2 blocking, among others, have improved the outcome of patients with TP53alt. As TP53 mutations may occur de novo during relapse, it is recommended that TP53alt assessment should be performed before initiating the first and every subsequent line of therapy.21
Immunoglobulin mutational statusAccording to the time when transformation took place, CLL B cells might be equivalent to normal cells that successfully passed through germinal centers resulting in a mutated phenotype (M-CLL). Alternatively, if they had not passed through germinal centers, CLL B-cells would show an unmutated (U-CLL) germline configuration.
The amount of somatic hypermutation (SHM) within the rearranged IG heavy variable (IGHV) genes of the clonotypic B-cell receptor (BcR) divides CLL in two large categories: one without or limited SHM (‘unmutated’ CLL, U-CLL) and another with significant SHM load (‘mutated’ CLL, M-CLL).
The mutational status of IGHV genes provides relevant prognostic information for CLL patients. CLL patients with high SHM show a longer overall survival than those with U-CLL.22 The IGHV mutational status is presently considered one of the most important prognostic factors to stratify CLL patients. For this reason, recommendations on how to perform and interpret IGHV mutational analysis in CLL have been recently intensively discussed to avoid discrepancies between laboratories.23
Discrimination between M-CLL and U-CLL is carry out by comparing a sequence from a CLL blood sample with an IGHV germline reference sequence. M-CLL is reported when a difference above 2% is found between a CLL IGHV rearrangement and the most identical germline VH gene sequence. Conversely, a CLL sample is classified as U-CLL when less than a 2% identity is found. In an alternative nomenclature, IGHV unmutated is the term used when 98% or more sequence homology to the nearest germline gene is found.24
In view of the prognostic relevance of the IGHV mutational status, specific ERIC recommendations are also proposed.24 IGHV mutational analysis optimization is performed at two levels, firstly by harmonizing technical protocols to generate a reliable IGH sequence and, secondly, by generating a proper clinical interpretation and report. Briefly, descriptions for appropriately initiating the processing of samples, mononuclear cells or purified B cells for DNA or cDNA analysis, design of IGHV leader primers or also available IGHV FR1 primers. Leader primers are considered to be the optimal choice followed by clonality testing revealed by heteroduplex analysis or Gene Scan / fragment analysis and direct sequencing protocols. Furthermore, interpretation and clinical reporting are also discussed. Finally, it is worth noticing that presence of an IGHV3-21 gene segment may be associated with an unfavorable prognosis independently of the IGHV mutational status.24,25
Minimal residual disease (MRD) has an important role in assessing CLL treatmentWhen the complex biology of CLL was revealed a large number of therapies were established. Therefore, defining suitable endpoints for evaluating response, relapse, refractoriness and resistance to therapy were strongly searched. Moreover, minimal residual disease eradication (<0.01% CLL cells) has become the novel surrogate endpoint for clinical studies.
MRD has been found to be an independent predictor of progression-free survival (PFS) and overall survival (OS) in CLL.26,27 Assessing MRD may improve the possibility of predicting the duration of PFS. Among CLL patients with low MRD, PFS did not differ significantly between complete response (CR) and partial response (PR). On the other hand, PFS was longer in patients with partial response (PR) with non-detectable MRD than for patients with CR with detectable MRD, showing that the MDR status may be more relevant for PFS than response rate.28
MRD as a surrogate indicator of treatment effectiveness may allow to evaluate the efficacy of new therapies without the need of continuous follow ups and the long time presently required for assessing PFS as endpoint. For instance, MRD > 0.010% (10−4) threshold is an independent predictor of PFS in CLL patients treated with chemo immunotherapy and also with new available therapies. Moreover, complete CLL B cell eradication with more sensitive procedures is highly expected because persistence of low levels of leukemic cells is a major cause of relapse.
Therapy response can be evaluated by different procedures and defined by different criteria with manifold levels of sensitivity. Definition of time points and technical tools are important parameters for proper monitoring of response. Different methodological approaches can be used for detecting MRD, like high-throughput sequencing (ClonoSEQ assay), allele-specific oligonucleotide PCR, and multiparametric flow Cytometry (MFC). This last procedure is rapid and reproducible, permitting quantitative estimates of CLL B cell decrease or disappearance. MFC analysis allows monitoring based on serial evaluations along treatment, becoming the strategy of choice for MRD detection in most centers.
Finally, although it is too early to conclude that MRD might be a valid surrogate endpoint, prospective studies investigating MRD role in CLL will most probably show its relevance as a surrogate CLL endpoint.
Conflicts of interestThe authors declare no conflicts of interest.