The para-Bombay phenotype, or H-deficient secretor, results from different mutations of the FUT1, with or without the FUT2 mutation. Consequently, there is an absent or weak expression of the H antigen on red blood cells (RBCs). Routine ABO blood grouping for two siblings with blood group O showed discrepant results with their parental blood group AB. Fragments encompassing the entire coding region of the FUT1 and FUT2 genes were investigated.
MethodsBlood and saliva specimens were collected to verify the correct ABO grouping by cell grouping, serum grouping and the hemagglutination inhibition (HI) test, respectively. The FUT1 and FUT2 genomes were identified using the whole-exome sequencing (WES) in two children's DNA blood specimens and may have caused, or been relative to, their blood group. Genetic variations of the FUT1 and FUT2 genes have been investigated in the other family members using the Sanger sequencing.
ResultsThe serologic reaction results of the proband revealed that A, B and H antigens were absent on RBCs, and that the serum contained anti-H. However, ABH and AH antigens were present in the saliva PB1 and PB2, respectively. The probands PB1 and PB2 were assigned as AB and A blood groups, respectively. Blood genotyping confirmed that heterozygous mutations of the FUT1 gene, c.551_552delAG, were identified. Three family members, PB3, PB, and PB8, also showed normal ABO blood groups, but their genotypes were also the FUT1 mutation c.551_552delAG.
ConclusionsThe FUT1 mutation c.551_552delAG may result in the reduced or absent H antigen production on RBCs, which characterizes the para-Bombay phenotypes. Blood genotyping is essential if these individuals need a blood transfusion or are planning to donate blood.
The ABO blood group was the first blood group system discovered by Karl Landsteiner in 1900. The ABO blood group is the most crucial blood group because it can produce antibodies against an antigen that is not naturally present in the individual (naturally occurring antibodies, or Nabs). The ABO-incompatible blood transfusion can lead to an acute hemolytic transfusion reaction (AHTR) and may result in hypotension, disseminated intravascular coagulation (DIC), renal failure and death. For this reason, accurate blood grouping is essential in the event of a blood transfusion.1,2
The ABO blood group expression is governed by 3 genes: the ABO gene on the chromosome 9q34, the FUT1 gene (H gene) and the FUT2 gene (secretor gene) located close to each other on the chromosome 19q13.3. Antigens of the ABO blood group are synthesized sequentially on the H antigen, which is the precursor of the A and B antigens. Based on the site of the antigen production, two mechanisms regulate H antigens. The FUT1 gene controls the first mechanism for fucosyltransferase production, adding the L-fucose sugar to type 2 chain oligosaccharides on the glycoprotein and glycolipid components on red blood cell surfaces and then synthesize the H antigen that has antigenic properties. The FUT2 gene controls the second mechanism for the fucosyltransferase production, adding the L-fucose sugar to type 1 chain oligosaccharides. These mechanisms form the H antigen in the secretions, such as saliva, tears, milk, sweat, semen, serum, cervical secretion and ascites fluid. H antigens on the cell surfaces and in the secretions are converted to the A/B antigen by the A/B transferase.1–4
The FUT1 and FUT2 gene mutation revealed rare blood groups, such as the production of the Bombay and para-Bombay phenotypes. The Bombay phenotype caused by the FUT1 and FUT 2 mutations indicated the homozygous nonfunctional H gene (hh) and homozygous nonfunctional secretor gene (sese) and, therefore, cannot synthesize fucosyltransferase for the production of the type 2 and type 1 H antigen. The Bombay blood group red blood cell does not contain the A, B and H antigens. Thus, there are naturally occurring antibodies, anti-A, B and H in serum, whereas in secretions, such as saliva, the A, B and H antigens are not detected.1,5,6
The para-Bombay phenotype is another rare blood group with an important characteristic, the H-deficient secretor. The H gene is not expressed on either side of the chromosome, but at least one secretor gene is expressed. Thus, the H antigen is absent on the red blood cell surface, but there may be a small amount of antigens
A or B, which is absorbed from A or B substances in the plasma in which the ABO and secretor gene are still functioning normally.1–3 The cause of the nonfunctional H gene is a mutation of the H gene, with more than 40 alleles that have been found to cause H-deficient phenotypes.2,3,6 The para-Bombay phenotype may also be an H-partially deficient non-secretor, with small amounts of the type 2 H antigen on the red blood cell surface, but without the type 1 H antigen in secretion.2
Since the para-Bombay phenotype has weakly expressed A or B antigens, the conventional cell grouping method may not detect the agglutination, even if the red blood cells have the antigen on the surface. Therefore, the results were inconsistent with the individual's serum grouping, resulting in the ABO discrepancy. Probable cause analysis and choosing the appropriate test for accurate blood group results, including selecting blood components to provide safe blood to patients, is paramount. Despite this, para-Bombay individuals may make a potent antibody against the H antigen (like those with the Bombay phenotype). As a result, they may require H negative blood for transfusion.1,2,4,5 Currently, more than 40 mutations on FUT1 alleles associated with either the Bombay or para-Bombay phenotypes have been described. The nucleotide substitutions, insertions and deletions of short DNA strands were reported. This study investigates the FUT1 and FUT2 genes from a sibling with group O who was born from group AB parents and nine other family members. The results were analyzed and compared with known mutations.
Materials and methodsSpecimen collectionThe reason for the suspected ABO incompatibility was that the father and mother are blood group AB and could transfer an A, B or AB allele to his son or daughter. This couple could have had blood group A, B or AB children, but this was not so. Their children are in the O blood group by the standard hemagglutination test. The parents of these children were deeply concerned and eager to know their children's real blood group, leading to a special test.
Two siblings' blood (big brother and little sister), blood and saliva specimens were collected to perform the correct ABO grouping. In addition, blood and saliva specimens of their family members, including father, mother, grandfather, two grandmothers, two father's siblings and two mother's siblings, were collected to determine the ABO grouping related to the children's blood group. Ethical approval for this study was obtained from the Human Research Ethics Committee of Walailak University, Thailand (WUEC-20-339-01).
ABO blood grouping and saliva testingAll eleven blood and saliva specimens were performed the ABO grouping at the blood bank laboratory, Maharaj Nakhon Si Thammarat Hospital, Thailand. The ABO blood grouping was typed by the serological method; Anti-A, Anti-B and Anti-H were used for cell grouping and A cells, B cells and O cells, for serum grouping. The ABH antigen in saliva was typed by the hemagglutination inhibition (HI) test with A cells, B cells and O cells. All antiserum and cell reagents were from the National Blood Centre Thai Red Cross Society, Bangkok, Thailand. The Anti-A, Anti-B and Anti-H were murine monoclonal antibodies (MoAb).
Whole-exome sequencing (WES)The two children's DNA blood specimens were further identified by the protein-coding regions of the genome that may cause, or be relative to, their blood group using the whole-exome sequencing (WES). The WES was manufactured by GENEWIZ, South Plainfield, NJ.
Sanger sequencingGenetic variations, such as single-nucleotide variants (SNVs) and Indels of the FUT1 and FUT2 genes from WES, have investigated the nucleotide sequence of DNA in all eleven specimens using the Sanger sequencing. The primer pair used for the Sanger sequencing is the FUT1 and FUT2 genes with 435 and 449 bp; 5’ GCCGGTTTGGTAATCAGATGG 3’ for the forward primer and 5’ AGCGCTGAGGCATAACCTG 3’ for the reverse primer of the FUT1 gene. For the FUT2 gene, 5’ GAGTACGCCACACTGTACG 3’ was used for the forward primer and 5’ AGGGCCTGCTGTAGGTATC 3’, for the reverse primer. The PCR condition was run for 1 minute at 95°C, 15 seconds at 95°C, 20 seconds at 60°C and 30 seconds at 72°C in 35 cycles. The Sanger sequencing was manufactured and analyzed by GENEWIZ, South Plainfield, NJ.
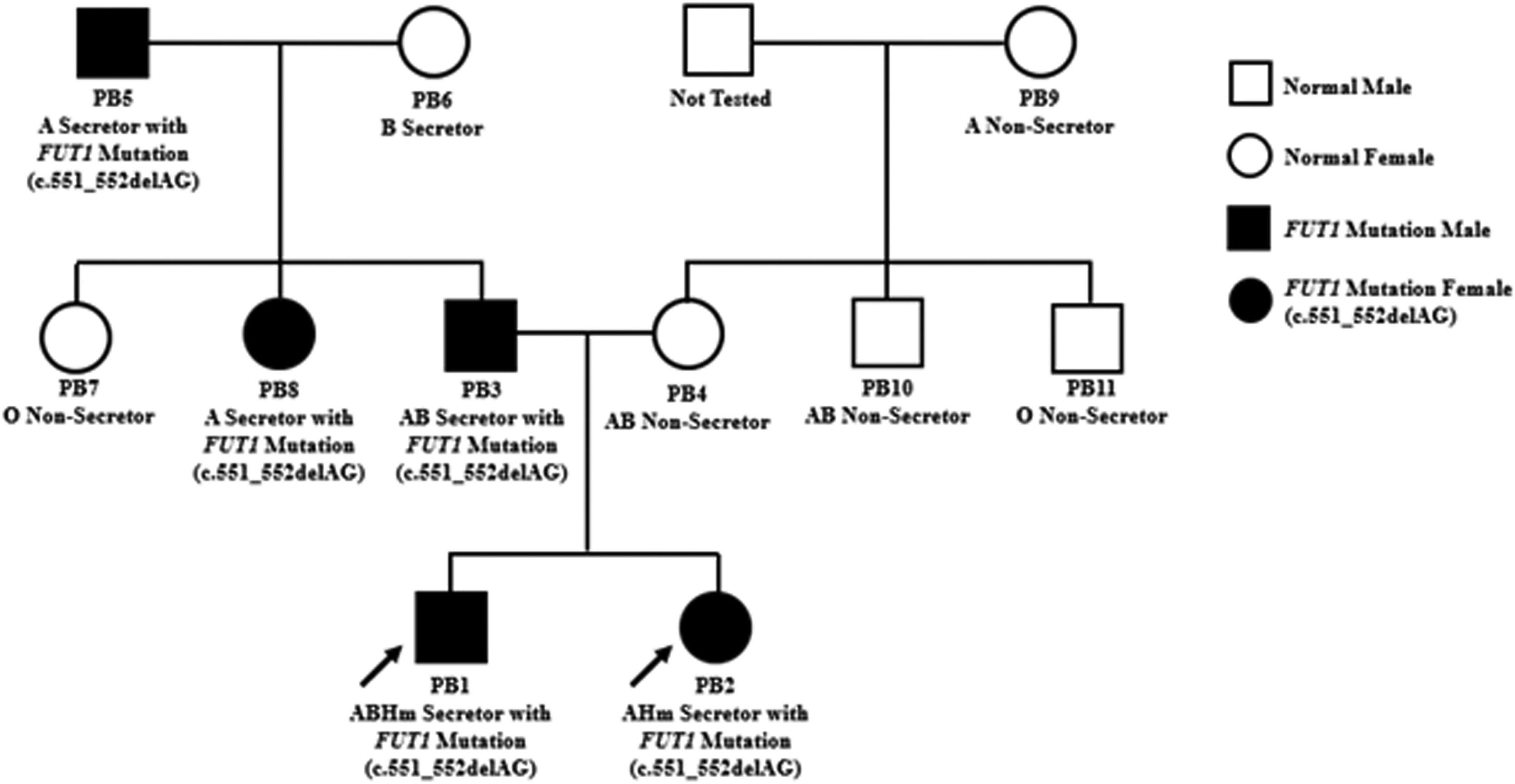
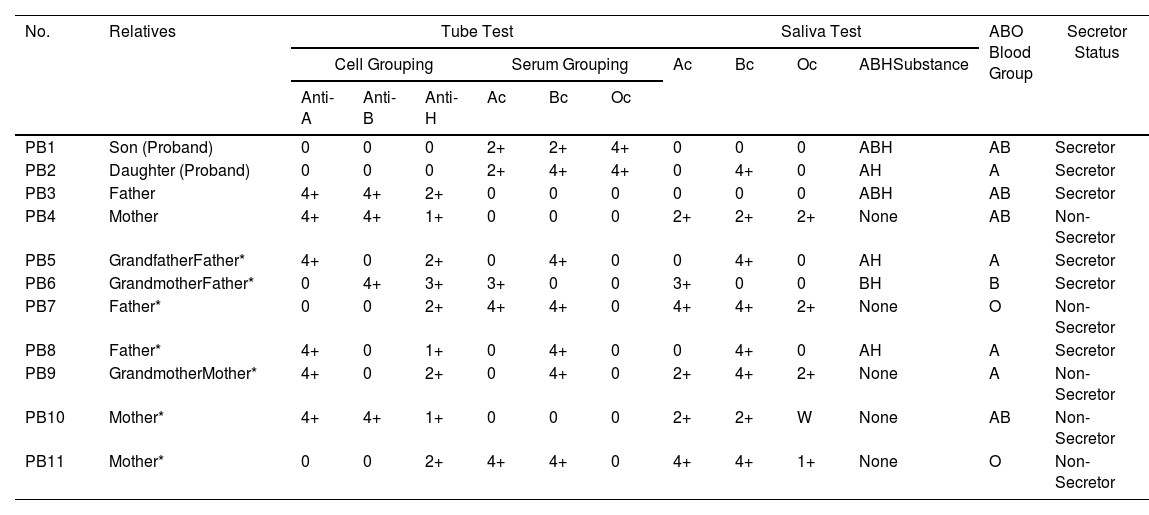
ResultsSerological results of the ABO blood group (tube test, cell grouping and serum grouping) and saliva test are shown in Table 1. Eleven specimens were tested to identify the ABO blood group and secretor status. The PB1 is a proband with the AB para-Bombay (ABHm) secretor and the PB2 is also a proband with the A para-Bombay (AHm) secretor. According to the heredity on the paternal side, the grandfather (PB5) had the A secretor and the grandmother (PB6), the B secretor. These grandparents spawned the PB3 (father) with the AB secretor, the PB7 (father's sibling) with the O non-secretor and the PB8 (father's sibling) with the A secretor. On the maternal side, the grandmother (PB9) had the
Serological results of ABO blood group and saliva test.
| No. | Relatives | Tube Test | Saliva Test | ABO Blood Group | Secretor Status | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cell Grouping | Serum Grouping | Ac | Bc | Oc | ABHSubstance | ||||||||
| Anti-A | Anti-B | Anti-H | Ac | Bc | Oc | ||||||||
| PB1 | Son (Proband) | 0 | 0 | 0 | 2+ | 2+ | 4+ | 0 | 0 | 0 | ABH | AB | Secretor |
| PB2 | Daughter (Proband) | 0 | 0 | 0 | 2+ | 4+ | 4+ | 0 | 4+ | 0 | AH | A | Secretor |
| PB3 | Father | 4+ | 4+ | 2+ | 0 | 0 | 0 | 0 | 0 | 0 | ABH | AB | Secretor |
| PB4 | Mother | 4+ | 4+ | 1+ | 0 | 0 | 0 | 2+ | 2+ | 2+ | None | AB | Non-Secretor |
| PB5 | GrandfatherFather* | 4+ | 0 | 2+ | 0 | 4+ | 0 | 0 | 4+ | 0 | AH | A | Secretor |
| PB6 | GrandmotherFather* | 0 | 4+ | 3+ | 3+ | 0 | 0 | 3+ | 0 | 0 | BH | B | Secretor |
| PB7 | Father* | 0 | 0 | 2+ | 4+ | 4+ | 0 | 4+ | 4+ | 2+ | None | O | Non-Secretor |
| PB8 | Father* | 4+ | 0 | 1+ | 0 | 4+ | 0 | 0 | 4+ | 0 | AH | A | Secretor |
| PB9 | GrandmotherMother* | 4+ | 0 | 2+ | 0 | 4+ | 0 | 2+ | 4+ | 2+ | None | A | Non-Secretor |
| PB10 | Mother* | 4+ | 4+ | 1+ | 0 | 0 | 0 | 2+ | 2+ | W | None | AB | Non-Secretor |
| PB11 | Mother* | 0 | 0 | 2+ | 4+ | 4+ | 0 | 4+ | 4+ | 1+ | None | O | Non-Secretor |
Note 1) Ac = A cells, Bc = B cells and Oc = O cells.
A non-secretor. She spawned the PB4 (mother) with the AB non-secretor, the PB10 (mother's sibling) with the AB non-secretor and the PB11 (mother's sibling) with the O non-secretor.
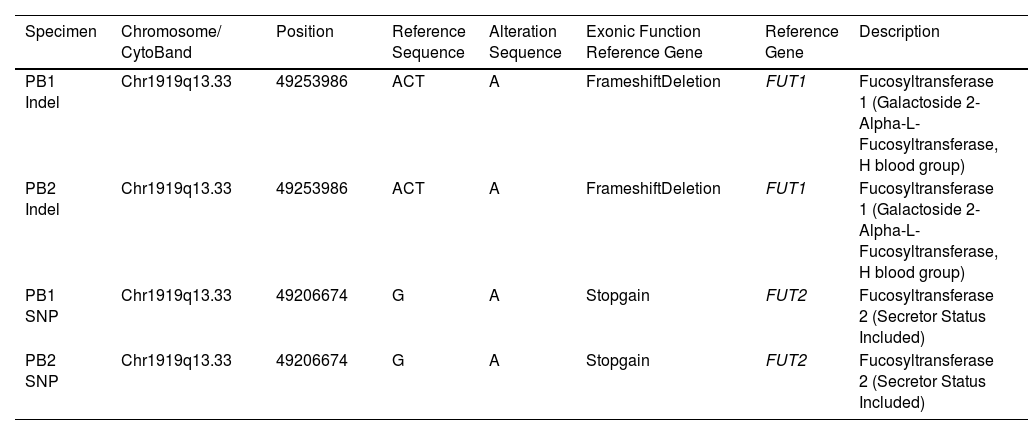
The PB1 and PB2 were further tested to identify the mutation of the FUT1 and FUT2 genes using whole-exome sequencing (WES) (Table 2).7,8 The results showed that the PB1 and PB2 had the indel 49253986 ACT>A on the FUT1 exon of the chromosome 19q13.33. If a mutation disrupts the reading frame, then the entire DNA sequence following the mutation will be misread. The WES results on DNA from the PB1 and PB2 showed homozygosity for the indel ACT>A at position 49253986 on chromosome 19q, which corresponded to the FUT1 c.551_552delAG (FUT*01N.06) and resulted in the p.Trp143Ter of the H-phenotype, and were further validated using the Sanger sequencing.
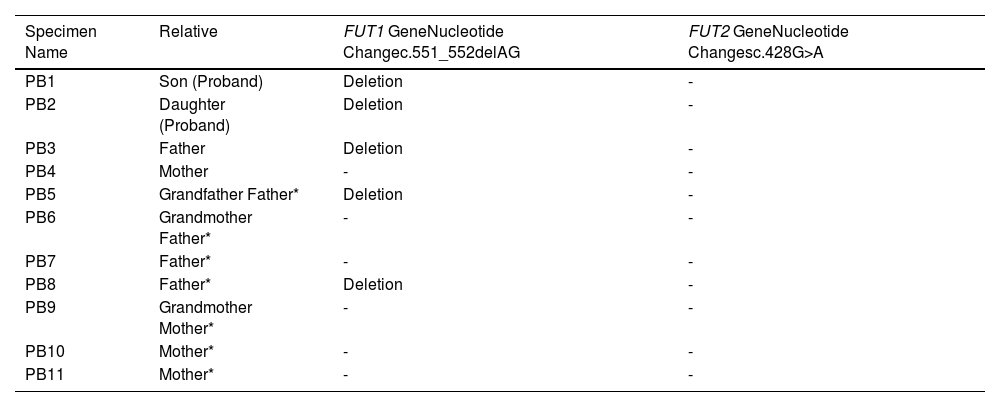
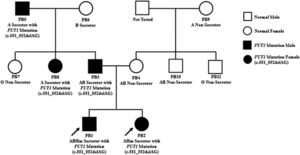
The validation results of the FUT1 indel 49253986 ACT>A causes the nucleotide change c.551_552delAG of both the FUT1_F and FUT1_R in PB1, PB2, PB3, PB5 and PB8. While the FUT2 single-nucleotide polymorphism (SNP) 49206674 G>A causes the nucleotide changes, the c.428G>A showed no change in either the FUT2_F or FUT2_R in all PB1 to PB11 specimens. (Table 3). Although the results of the WES showed the heterozygous SNP 49206674 G>A on the FUT2 exon of chromosome 19q13.33 in PB1 and PB2, Sanger sequencing did not. This may be caused by de novo or mosaic de novo mutation less than 20%that cannot be detected by Sanger sequencing.9 Such false-positive events from the WES in PB1 and PB2 may be caused by misaligning sequencing reads to a reference sequence (RefSeq). Moreover, inaccuracies or biases of the RefSeq, compared to a specific local population, are other sources of false-positive genotype calls in next-generation sequencing (NGS) data.10 Misalignments of short-length sequencing read to a reference sequence are influenced by seed-based strategies or algorithms for complete alignment permutations.11-13 These problems often arise in low-complexity regions14 or result from misaligning multiple copies of genes, paralogues or pseudogenes.15 Thus, the WES may be unreliable for detecting copy-number variations, almost all of which extend beyond the targeted regions. Finally, Sanger sequencing is a powerful tool in detecting potential disease-causing mutations within the WES regions, particularly those due to SNVs. Another event that the SNP 49206674 G>A on the FUT2 exon of chromosome 19q13.33 in PB1 and PB2 did not detect by Sanger sequencing may be caused by heterozygous mutation with incomplete penetrance. Indeed, blood groups, including the para-Bombay, are inherited by children (PB1 and PB2) from their parents (PB3 and PB4). This study showed the FUT1 nucleotide change c.551_552delAG from the paternal inheritance, but no FUT1 mutations were found in the maternal inheritance. The author expected that mutation of the FUT1 is passed on to the children's blood group, possibly from the mother, with a small amount of mutation or de novo mutation that Sanger sequencing cannot detect. The family pedigree represents the inheritance of FUT1 genes. The father's side consists of the grandfather (PB5), grandmother (PB6) and their one son and two daughters (PB3, PB7 and PB8, respectively) and one son is the proband's father (PB3). The mother's side consists of a grandmother (PB9) and their one daughter and two sons (PB4, PB10 and PB11, respectively) and one daughter is the proband's mother (PB4). PB3 and PB4 have their two probands (PB1 and PB2). The FUT1 mutation c.551_552delAG was detected in PB1, PB2, PB3, PB5 and PB8, while PB4, PB6, PB7, PB9, PB10 and PB11 were not (Figure 1).
Validation results of FUT1 indel and FUT2 SNP in the PB1 to PB11 using Sanger sequencing.
Note 1) * = Father's or Mother's Relative or Sibling 2) - = No Change.
The pedigree concluded that there were two members in the para-Bombay blood group in this family, PB1 and PB2 (proband). The FUT1 mutation associated with the expression of the para-Bombay phenotype was found among 11 family members, five of which were found to be mutated in the FUT1 c.551_552delAG, the PB1 (proband), PB2 (proband), PB3 (father), PB5 (father's sibling) and PB8 (grandfather). Therefore, it is possible that a mutation of the FUT1 from the grandfather was passed on to the father and father's sibling and became a para-Bombay phenotype in both children. The PB1 and PB2 were identified as para-Bombay phenotypes from other samples in which the FUT1 mutation c.551_552delAG inherited from their father was detected. This could be due to a very low mutation in the FUT1 inherited from the mother and could not be detected by this method.
Due to the discrepant results from routine ABO blood group typing for pre-transfusion testing, the FUT1 mutation c.551_552delAG has been reported using sequencing analysis,15 but not reported in the Thai population. Since the anti-H antiserum is not commonly used for the ABO grouping, the para-Bombay phenotype cases would go unnoticed. Although very rare, in particular, when examining individuals exhibiting an unusual ABO blood group, the presence of an H-deficient phenotype should be considered. In such phenotypes, particular attention should be paid to the possibly mistaken reverse grouping results for the safety of blood transfusions.16
Mutations that inactivate the FUT1 can result in the para-Bombay phenotype where no A, B or H antigen is produced on the RBC.17 The H blood group system consists of one antigen H that is carried on glycolipids and glycoproteins on the RBC membrane, where the fucosyltransferase product of the FUT1 synthesizes it and on glycoproteins on epithelial cells and in body fluids, where the fucosyltransferase product of the FUT2 synthesizes it. In group O individuals, the H antigen is the terminal antigen. However, the H antigen serves as the precursor structure for A and B blood group-specific glycosyltransferases in group A and B individuals. Thus, group O people will test H+ strongly, whereas groups A, B and AB will express very little H antigen. Mutations that negatively affect the α2FucT1 enzyme activity (encoded by the FUT1 will result in reduced or absent H production (and a concomitant decrease in A or B antigens in individuals in whom those enzymes are encoded). A total absence of H, A and B antigens is called the Oh or Bombay phenotype. Weak expression is referred to as the para-Bombay phenotype. The enzymes α2FucT1 and α2FucT2 are single-pass type II membrane glycoproteins in the Golgi. The α2FucT1 protein contains 365 amino acids encoded by the FUT1 or H, if the analysis predicts a blood group antigen. The FUT2 produces two transcripts, one of 343 amino acids and another, which is more abundant, of 332 amino acids. The longer transcript encodes a protein with approximately one-fourth of the enzymatic activity and the shorter form is considered the active enzyme. Thus, the numbers given below are counted from the second initiating codon. The FUT2, or Se, encodes the α2FucT2 protein, if the analysis is to predict a blood group antigen.
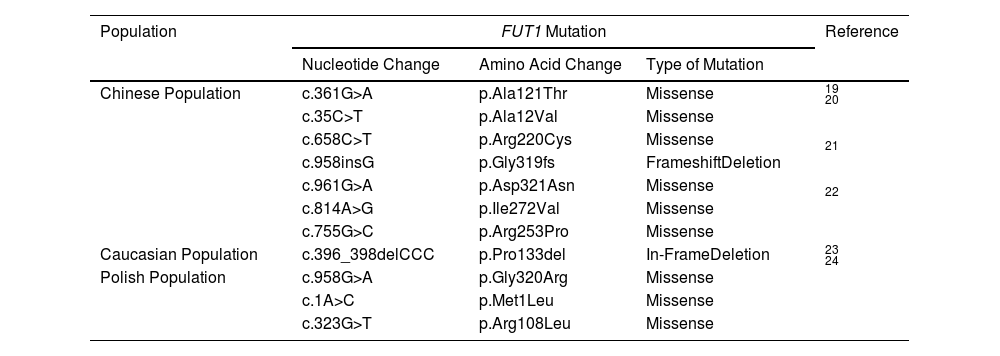
Molecular mutagenicity studies have not been reported in the para-Bombay blood group in Thailand. Still, in different population groups, Table 4 shows a novel FUT1 allele (FUT1*c.361G>A, summarized and identified in a Chinese individual with the para-Bombay B phenotype. The FUT1 c.361G>A mutation may significantly downregulate the expression of H antigens on RBCs by damaging the enzyme conformation.18 The para-Bombay phenotype and its variants would be explored further in performing a simple saliva secretor test, with the use of the anti-H lectin in blood grouping. Hence, it should be borne in mind that a thorough analysis of any blood group discrepancy is warranted, as it has significant clinical implications.19 Mutations of the FUT1 were discovered: the h(35T+658T)/h(35T+658T) is responsible for the para-Bombay phenotype of the propositus. The genotype is rare even in Chinese para-Bombay populations.20 In a Chinese para-Bombay individual, novel FUT1 mutations have been identified in the proband's FUT1 gene, homozygous for c.958insG, c.961G > A, c.814A>G and c.755G>C.21,22 Furthermore, an H+w individual was homozygous for a base triplet deletion (c.396_398delCCC), leading to a single amino acid (p.Pro133del) in the fucosyltransferase 1.23 In addition, in the Polish population, the DNA sequence analysis of the coding region of the FUT1 revealed a different and previously unpublished mutation in each of the three donors: 1) homozygous for c.958G>A, which encodes a change of p.Gly320Arg; 2) homozygosity for c.1A>C, which encodes a change of the p.Met1Leu but, more importantly, disrupts the translation-initiating start codon for the FUT1 coding region, and; 3), homozygous for c.323G>T, a change that predicts the p.Arg108Leu.24
FUT1 Variants leading to the Para-Bombay phenotype in different populations.
| Population | FUT1 Mutation | Reference | ||
|---|---|---|---|---|
| Nucleotide Change | Amino Acid Change | Type of Mutation | ||
| Chinese Population | c.361G>A | p.Ala121Thr | Missense | 19 |
| c.35C>T | p.Ala12Val | Missense | 20 | |
| c.658C>T | p.Arg220Cys | Missense | ||
| c.958insG | p.Gly319fs | FrameshiftDeletion | 21 | |
| c.961G>A | p.Asp321Asn | Missense | ||
| c.814A>G | p.Ile272Val | Missense | 22 | |
| c.755G>C | p.Arg253Pro | Missense | ||
| Caucasian Population | c.396_398delCCC | p.Pro133del | In-FrameDeletion | 23 |
| Polish Population | c.958G>A | p.Gly320Arg | Missense | 24 |
| c.1A>C | p.Met1Leu | Missense | ||
| c.323G>T | p.Arg108Leu | Missense | ||
The FUT1 mutation c.551_552delAG may result in reduced or absent H antigen production on RBCs, which characterizes the para-Bombay phenotypes. Since anti-H antiserum is not commonly used for ABO grouping, para-Bombay phenotype cases would go unnoticed. It was possible to make this finding for two siblings of the para-Bombay blood group. Blood genotyping is essential if they need a blood transfusion or are planning to donate blood.
FundingThis study was supported by the Individual Research Grant from Walailak University (grant number WU-IRG-64-002).
Authorship contributionsYR and TC conceived the study. YR and SN determined the ABO grouping, saliva testing, whole-exome sequencing and Sanger sequencing. YR and TC analyzed all the data. YR wrote the manuscript. NC and TC critically revised the manuscript. All the authors approved the final version of the manuscript.
Funding statementThis study was supported by the Individual Research Grant from Walailak University (grant number WU-IRG-64-002).
Ethics approval statementEthical approval for this study was obtained from the Human Research Ethics Committee of Walailak University, Thailand (WUEC-20-339-01).