The BCR-ABL1 oncogene is the molecular hallmark of chronic myeloid leukemia (CML) and Philadelphia chromosome-positive acute lymphoblastic leukemia (Ph+ ALL). The most common BCR-ABL1 transcript in Ph+ ALL patients results from the fusion of BCR exon 1 to ABL1 exon a2 (e1a2) and is present in approximately two thirds of adults cases. Remaining patients express a fusion between BCR exon 13 or 14 and ABL1 exon a2 (e13a2 and e14a2 respectively) however a minority express atypical transcripts usually due to splicing of alternative BCR or ABL1 exons. These variants become apparent upon discordant diagnostic cytogenetic and molecular testing and may be resolved by multiplex, reverse transcription (RT)-PCR or sequencing approaches.1 In a recent international survey of BCR-ABL1 transcript types in more than 34,000 CML patients, 1.93% of cases expressed these atypical forms yet their frequency in adult Ph+ ALL is not widely appreciated.2 The incidence and type of BCR-ABL1 transcripts in national cohort of adult Ph+ ALL patients were considered in order to inform appropriate, prospective diagnostic and measurable residual disease (MRD) monitoring approaches.
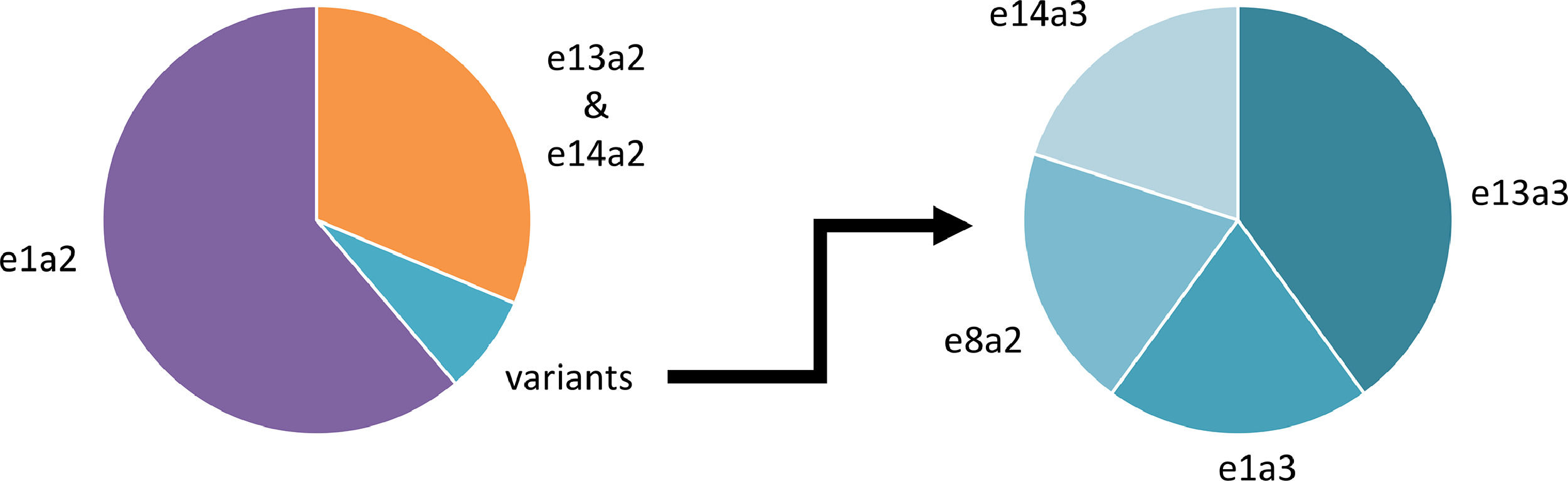
Ph+ ALL patients were identified from January 2005 to December 2020 inclusive at a national center for the molecular diagnosis and monitoring of BCR-ABL1-positive leukemias. All patients had cytogenetic or fluorescence in situ hybridization confirmation of a t(9;22) translocation or BCR-ABL1 fusion signal respectively. BCR-ABL1 detection was performed using a standardized RT-PCR approach throughout this period with Sanger sequencing used for confirmation of an atypical fusion transcript.3 Of 67 Ph+ ALL patients identified, typical e1a2 BCR-ABL1 transcripts were detected in 41 (61.2%; median age 43 years, range 18–79), e13a2 or e14a2 in 21 (31.3%; median age 51 years, range 22–70) and atypical transcripts in five (7.5%; median age 58 years, range 20–72) (Figure 1).
The above demonstrates that the frequency of atypical BCR-ABL1 transcripts in this adult Ph+ ALL population appears higher than that previously reported though this may be a reflection of the cohort size.1 The clinical importance of precise identification of the BCR-ABL1 transcripts is twofold. Monitoring of MRD by quantification of BCR-ABL1 transcripts is now an integral part of Ph+ ALL patient management with significant advances made towards harmonization of methodological approaches.4,5 Therefore, characterization of a variant transcript type allows the selection of correct primers and probes for quantitative RT-PCR or digital PCR. To date, there is limited information on MRD profiles in those Ph+ ALL patients expressing atypical BCR-ABL1 transcripts. Secondly, there is accumulating evidence for a BCR-ABL1 genotype-tyrosine kinase inhibitor (TKI) response correlation in patients with CML but a similar relationship in Ph+ ALL patients is unknown.6 Some evidence suggests that Ph+ ALL patients expressing e13a2 or e14a2 transcripts experience inferior outcomes when compared to their e1a2 BCR-ABL1 counterparts which is in contrast to that found in CML.7,8 Therefore, knowledge of the transcript type might possibly inform selection of chemotherapy and TKI. With improvements in the overall survival of adult patients with Ph+ ALL, consensus MRD methodologies are increasingly required for those patients expressing the variant BCR-ABL1 transcript types.
Ethical disclosuresNo funding was received for this work. This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors. This study was performed as part of routine standard of care in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki declaration and its later amendments. Informed consent was obtained from participants at the referring centres. All authors contributed to data acquisition, manuscript preparation and approved the final version.