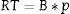Alloimmunization is a major problem in transfusion practice due to the clinical complications of the patients and the difficulty of choosing a unit of compatible blood product. Serological methods are widely used in blood banks, but they not always determine the phenotype. Thus, genotyping is an important complement to the serology tool as it allows one to predict the phenotype from deoxyribonucleic acid (DNA) with high accuracy.
ObjectiveTo compare the centrifugation gel, microarray, Restriction Fragment Length Polymorphismone PCR (PCR-RFLP) and Sequence-Specific Primer PCR (PCR-SSP) techniques, in terms of cost, reaction time and reliability of the results.
MethodsThe RHCE, Kidd, Kell and Duffy blood group systems were chosen to determine the approximate cost of each technique, considering the reagents used in both methods and considering only one sample. The time required for the development of each reaction was obtained at the Maringa Regional Blood Center and Immunogenetics Laboratory at the State University of Maringa. Data from Microarray reactions were obtained at the Campinas Blood Center. The results of phenotyping and genotyping of the 16 samples were compiled in a spreadsheet and compared.
ResultsThe PCR-SSP was more economical compared to other methods, and the serological method was faster than the molecular methods. However, all methods proved to be effective and safe in the detection of erythrocyte antigens.
ConclusionAnalyzing the advantages and limitations of the molecular and serological methods tested in this study, we note that both are important and complementary. However, the choice of a methodology depends on the reality and needs of each health service.
The detection of blood group antigens is essential in transfusion practice in order to prevent alloimmunization, especially in multiple-transfused patients. Erythrocyte antibodies are a clinically significant problem in transfusion medicine because they can lead to acute transfusion reactions and hemolytic disease of the fetus and newborn, increasing the morbidity and mortality rate of the patients. In addition, alloimmunization may delay the localization of a compatible blood bag.1–3 The probability of an individual producing one or more anti-erythrocyte antibodies is approximately 1% per unit of blood transfused, and in chronically transfused patients the alloimmunization rate may reach 50%.4–6
Blood group systems are characterized by the presence or absence of antigens on the surface of erythrocytes.5 Currently, 36 red blood cell systems have been described.7 In addition to the ABO and Rh compatibility for prevention of hemolytic reactions, other immunogenic blood group antigens may be considered, especially C, c, E, and e of the Rh and the Kell, Kidd, Duffy and MNS systems.8
Several methodologies for the detection of erythrocyte antigens are known. The classical method is phenotyping, currently performed by hemagglutination tests, which detect the gene product by binding antisera to specific antigens. The limitation of serological tests is to find specific antibodies for rare erythrocyte antigens.9 Besides, they are not always able to determine the phenotype due to lower antigen expression on the erythrocyte surface, or yet recent transfusions (<3 months), and certain pathologies, such as autoimmune hemolytic anemia and aplastic anemia.10
Advances in molecular biology have made it possible to implement and improve new methodologies to increase the safety of blood transfusion. Genotyping has been shown to be effective and advantageous in relation to phenotyping in some cases, as it provides means to predict the phenotype from genomic DNA with a high degree of precision.11,12 Several applications of molecular methods for erythrocyte genotyping have been described, among them being fetal DNA typing, determination of the blood group of a recently transfused patient, screening of blood donors to find phenotypes of rare blood groups, determination of the frequency of blood group polymorphisms in a population and blood typing of patients with autoimmune hemolytic anemia and donors for alloimmunized patients.12,13
The molecular basis for most erythrocyte antigens is known and numerous DNA analysis methodologies have been developed, all based on PCR. Single nucleotide polymorphisms (SNPs) can be determined by a variety of genotyping methods, including the sequence-specific primer PCR (PCR-SSP) using restriction enzymes (PCR-RFLP) and real-time PCR based on allele-specific extension and sequencing. It can also associate the PCR-SSP technique to the multiplex to facilitate the detection of several alleles simultaneously, as long as the alleles studied have products of different sizes. The most modern technique for DNA analysis is the Microarray, a method for large-scale erythrocyte genotyping.9,14,15
The PCR-SSP uses specific primers to detect the nucleotide sequence of the polymorphic alleles.16,17 The PCR-RFLP is characterized by the cleaving of the amplicons by specific endonucleases generating restriction profiles. Microarray genotyping is based on the detection of a color spectrum generated by contacting the target DNA with fluorescently labeled oligonucleotide probes deposited on a plate by an automated system which provides the results in graphs or tables.14,18
Considering the advantages and disadvantages of each method available for the detection of erythrocyte antigens and aiming at the minimization of costs and the agility of the reports produced without the loss of the safety procedure, the objective of this study was to compare the centrifugation hemagglutination gel test, PCR-RFLP, Microarray and PCR-SSP techniques, regarding the cost, reaction time and reliability of the results.
Material and methodsThis study was approved and conducted according to the norms recommended by the Ethics Committee on Human Research of the Maringa State University (CAAE: 0402.0.093.000-11).
The studied techniques were compared for the antigens determination of the RHCE (RHCE*E, RHCE*e), Kell (KEL*01, KEL*02), Duffy (FY*01, FY*02) and Kidd (JK*01, JK*02) systems, because these were tested by all techniques under analysis.
For the development of all genotyping methodologies (PCR-RFLP, PCR-SSP and Microarray) a first step of DNA extraction is required, which was performed by the QIAamp® DNA Blood Mini Kit, (Qiagen, Valencia, CA, USA), according to the recommendations of the manufacturer.
PCR-SSP genotyping was performed according to Liu et al.,16 with changes in cycling temperatures and MgCl2 concentration. The concentration of MgCl2 was 1.5mM for RHCE*E, KEL*01 and KEL*02, 1.0mM for FY*01 and FY*02 and 0.8mM for JK*01, JK*02 and RHCE*e. The proposed cycling conditions for these alleles were an initial denaturation step at 94°C for 2min, followed by 30 cycles of 30s denaturation at 94°C, 1min annealing at 72°C and 30s extension at 72°C, and a final elongation step was applied at 72°C for 5min. PCR-RFLP was performed according to protocols previously described in the literature,19–21 with some modifications.22 The HEA beadChipTM (Human Erythrocyte Antigen), Microarray platform (BioArray Solutions Ltd., Warren, NJ, USA) was used for large-scale genotyping. Phenotyping was performed by gel centrifugation method according to the recommendations of the manufacturer (DiaMed Latino América S. A., Lagoa Santa, MG, Brazil).
Phenotyping was performed at the Blood Distribution Department of the Maringa Regional Blood Center, the PCRs at the Immunogenetics Laboratory of the Maringa State University (LIG-UEM) and Microarray at the Molecular Biology Laboratory of the Hematology and Hemotherapy Center at Campinas State University (UNICAMP).
Cost analysisAfter estimating the reagents used for the methods under study, estimates from as many as three different suppliers were requested. In order to determine the gross value of each reagent, the average value obtained from the estimates was used. Common costs to all methodologies such as water, gloves, tips, pipettes and microtubes, as well as indirect costs, such as equipment, electricity and employees were not taken into account.
To calculate the cost with each reagent, the following equation was used:
RT represents the reagent cost and was proportional to the quantity used for one patient typing; B is the gross value of each reagent; and p is the value relative to the percentage of use of each reagent. The sum of all reagents costs used for typing one patient represents the total variable cost for each method.
For the genotyping techniques, the values of positive, negative and blank controls (without DNA) were included in the total cost, in addition to the sample.
Counting the time of the reactionTo determine the time required for the development of the technique, phenotyping was monitored at the Maringa Regional Blood Center according to Standard Operating Procedures, and the PCR-RFLP and PCR-SSP techniques, at the LIG-UEM. The microarray reaction time was obtained through information provided directly by the Molecular Biology Laboratory of the Hematology and Hemotherapy Center at UNICAMP. The total time to perform each technique was determined by the sum of each step time.
Analysis and comparison of resultsIn order to ensure the results obtained by the different methods, the selection of the samples, which had previously been phenotyped or genotyped at other hematology centers, was performed. Thus, rare, null and low expression phenotypes were not included. Phenotyping and Microarray results were obtained from the Francisco Beltrão Hemonucleo, Hemepar, Paraná and Campinas Blood Centers, Unicamp, Campinas, São Paulo, respectively, and the results of the PCR-RFLP and PCR-SSP were obtained in projects previously developed at the LIG-UEM. We selected 16 samples genotyped by the PCR-SSP. Among them, eight samples were also typed by the PCR-RFLP and serology and another eight, by Microarray. The results of all selected samples were compiled on a spreadsheet and compared for validation.
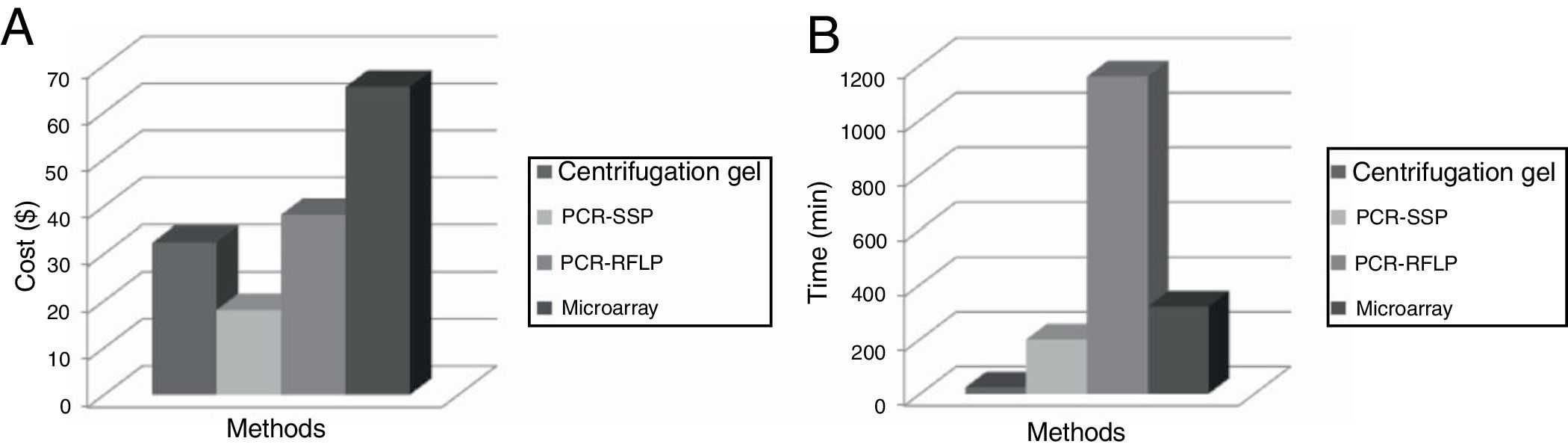
ResultsCost and time analysis of each reactionPhenotyping methodPhenotyping of RHCE, Kell, Duffy and Kidd erythrocyte antigens was performed by gel centrifugation methodology and the development time was approximately 25min (Figure 1). The reagents used and their respective costs are described in Table 1. The total variable cost was approximately U$32.45 per sample.
Approximate cost of the centrifugation gel technique for phenotyping one sample for the antigens from RHCE, Kell, Kidd and Duffy blood group systems.
| Reagents | Total cost (U$) | Cost/sample (U$) |
|---|---|---|
| ID-Perfil III-M-N-S-s-Fya-Fyb-ctl (1×12) | 56.96 | 4.74 |
| ID-Soro Teste Perfil III M-N-S-s-Fya-Fyb-ctl (6×1.4mL) | 238.39 | 8.51 |
| ID-Perfil II-k-Kpa-Kpb-Jka-Jkb-ctl (1×12) | 135.04 | 11.25 |
| ID Diaclon RH Subgroups+K (4×12) | 349.74 | 7.29 |
| ID-Diluent1 (2×100mL) | 85.65 | 0.43 |
| ID-Diluent2 (2×100mL) | 46.11 | 0.23 |
| Total | 911.89 | 32.45 |
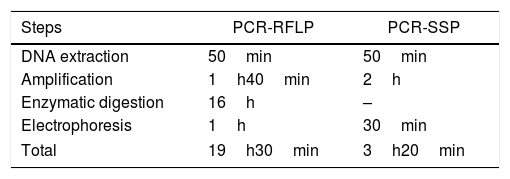
The PCR-RFLP technique requires 4 steps: DNA extraction, amplification, restriction enzyme digestion and electrophoresis. The time required for the performance of the technique was 19h30min. The PCR-SSP technique was performed in 3h20min (Table 2).
Time for PCR-RFLP and the PCR-SSP genotyping of the one sample for RHCE, Kell, Kidd and Duffy blood group systems.
| Steps | PCR-RFLP | PCR-SSP |
|---|---|---|
| DNA extraction | 50min | 50min |
| Amplification | 1h40min | 2h |
| Enzymatic digestion | 16h | – |
| Electrophoresis | 1h | 30min |
| Total | 19h30min | 3h20min |
DNA: deoxyribonucleic acid, PCR-RFLP: Polymerase chain reaction – Restriction Fragment Length Polymorphism, PCR-SSP: Polymerase chain reaction – Sequence Specific Primer.
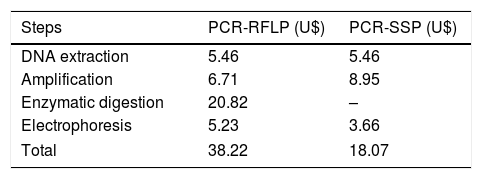
The total cost for the performance of the PCR-RFLP was higher than for the PCR-SSP (Table 3). The amplification cost includes the values of the Kit containing Taq DNA polymerase+MgCl2+Buffer, dNTP and primers. For the PCR-RFLP, the cost of digestion was relative to the price of the kit containing the restriction enzyme and buffer, proportional to the amount used. The cost of electrophoresis results from the values of agarose, 1× TBE (TRIS Base+boric acid+EDTA) and gel DNA stain.
Approximate cost of PCR-SSP and PCR-RFLP genotyping of one sample RHCE, Kell, Kidd and Duffy blood group systems.
| Steps | PCR-RFLP (U$) | PCR-SSP (U$) |
|---|---|---|
| DNA extraction | 5.46 | 5.46 |
| Amplification | 6.71 | 8.95 |
| Enzymatic digestion | 20.82 | – |
| Electrophoresis | 5.23 | 3.66 |
| Total | 38.22 | 18.07 |
PCR-RFLP: Polymerase chain reaction – Restriction Fragment Length Polymorphism, PCR-SSP: Polymerase chain reaction – Sequence Specific Primer.
The Microarray HEA beadChipTM (Human Erythrocyte Antigen) platform (BioArray Solutions Ltd., Warren, NJ, USA) uses a kit containing all reagents required to perform the technique. According to the data provided, a kit containing 12 blades has a cost of U$5760.00. Each blade allows the genotyping of 8 samples, so the approximate cost per sample was U$60.00, which added to the value of DNA extraction (U$5.46), generates a total cost of U$65.46. The average time to perform the technique is approximately 5h20min, according to Figure 2.
Figure 3 shows the cost and the time necessary to perform the identification of the antigens of the RHCE, Kell, Kidd and Duffy blood group systems for each methodology (phenotyping, PCR-SSP, PCR-RFLP and Microarray).
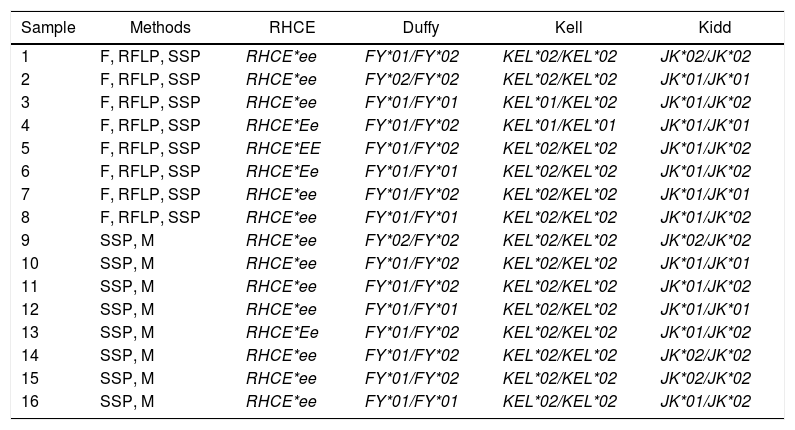
Analysis and comparison of resultsSamples were analyzed by at least two methods. Samples 1 through 8 were phenotyped by gel hemagglutination and genotyped by the PCR-RFLP and PCR-SSP and samples from 9 to 16 were genotyped by the PCR-SSP and Microarray (Table 4). The results are shown as a genotype nomenclature.18 No discrepancy was found among all analyzed samples.
Methods and genotyping definition for antigens of the RHCE, Duffy, Kidd and Kell blood group systems.
| Sample | Methods | RHCE | Duffy | Kell | Kidd |
|---|---|---|---|---|---|
| 1 | F, RFLP, SSP | RHCE*ee | FY*01/FY*02 | KEL*02/KEL*02 | JK*02/JK*02 |
| 2 | F, RFLP, SSP | RHCE*ee | FY*02/FY*02 | KEL*02/KEL*02 | JK*01/JK*01 |
| 3 | F, RFLP, SSP | RHCE*ee | FY*01/FY*01 | KEL*01/KEL*02 | JK*01/JK*02 |
| 4 | F, RFLP, SSP | RHCE*Ee | FY*01/FY*02 | KEL*01/KEL*01 | JK*01/JK*01 |
| 5 | F, RFLP, SSP | RHCE*EE | FY*01/FY*02 | KEL*02/KEL*02 | JK*01/JK*02 |
| 6 | F, RFLP, SSP | RHCE*Ee | FY*01/FY*01 | KEL*02/KEL*02 | JK*01/JK*02 |
| 7 | F, RFLP, SSP | RHCE*ee | FY*01/FY*02 | KEL*02/KEL*02 | JK*01/JK*01 |
| 8 | F, RFLP, SSP | RHCE*ee | FY*01/FY*01 | KEL*02/KEL*02 | JK*01/JK*02 |
| 9 | SSP, M | RHCE*ee | FY*02/FY*02 | KEL*02/KEL*02 | JK*02/JK*02 |
| 10 | SSP, M | RHCE*ee | FY*01/FY*02 | KEL*02/KEL*02 | JK*01/JK*01 |
| 11 | SSP, M | RHCE*ee | FY*01/FY*02 | KEL*02/KEL*02 | JK*01/JK*02 |
| 12 | SSP, M | RHCE*ee | FY*01/FY*01 | KEL*02/KEL*02 | JK*01/JK*01 |
| 13 | SSP, M | RHCE*Ee | FY*01/FY*02 | KEL*02/KEL*02 | JK*01/JK*02 |
| 14 | SSP, M | RHCE*ee | FY*01/FY*02 | KEL*02/KEL*02 | JK*02/JK*02 |
| 15 | SSP, M | RHCE*ee | FY*01/FY*02 | KEL*02/KEL*02 | JK*02/JK*02 |
| 16 | SSP, M | RHCE*ee | FY*01/FY*01 | KEL*02/KEL*02 | JK*01/JK*02 |
F: Phenotyping; RFLP: Polymerase chain reaction – Restriction Fragment Length Polymorphism; SSP: Sequence Specific Primer; M: Microarray.
The molecular genotyping of blood group antigens is an important aspect and is being introduced successfully in transfusion medicine. Low, medium and high-throughput techniques were developed for this purpose. Depending on the requirement of the center, such as screening for antigens of high or low prevalence where antisera are not available, correct typing of multiple-transfused patients, screening of antigen-negative donor units to reduce the rate of alloimmunization, a suitable technique can be selected.14
In this study we compared the hemagglutination by the gel centrifugation methodology and the techniques of genotyping to obtain the phenotype or genotype profile of a sample for the RHCE*E, RHCE*e, KEL*01, KEL*02, JK*01, JK*02, FY*01 and FY*02 alleles.
When the duration of each technique was compared, the results showed that the immunophenotyping required the shortest time to supply the results. Although the PCR-SSP used two independent amplification reactions, one for each allele, which generates more expense and greater preparation time for the reactions, it was reproduced in less time than the other genotyping methods. Even though the PCR-RFLP allows for the amplification of both alleles with a single pair of primers, that is, in a single reaction, the enzymatic digestion (overnight) considerably increases its reaction time.
In the total cost survey, the PCR-SSP reaction had the lowest cost, when compared to the other methodologies. Certainly, what led to the PCR-RFLP having a higher cost in relation to the PCR-SSP, as well as what was observed in the duration of the technique, was mainly the digestion step, due to the high cost of the restriction enzymes. It is important to note that for PCR reactions it is possible to reduce the cost considerably by processing more than one sample at a time, since in the processing of one or more samples the same controls can be used. Therefore, it is advisable to process a larger number of samples per reaction.
Despite the high cost, due to the use of advanced laboratory resources, the Microarray technique allows for large-scale genotyping, reducing the number of isolated procedures and consequently decreasing errors, permitting the identification of more than 30 different antigens,23 factors that must be considered in the analysis of cost-effectiveness.
In spite of the small number of samples used to compare the results among the different methodologies, all were shown to be efficient and reliable, since the results obtained were identical for all the samples. As the samples used in this study are from blood donors, we did not have any case of inconclusive phenotyping, an important factor in the evaluation of the sensitivity and specificity of this methodology.
Although the molecular methods are considered more sensitive and specific, we must consider that the presence of a particular gene does not guarantee its expression in the erythrocyte membrane, that is, patients with null phenotypes can be phenotyped as positive for a certain antigen.24 The phenotype, although faster, also presents some important limitations, such as the lack of antisera of rare antigens and the high cost for the acquisition of rare sera.9 Another difficulty is the phenotyping of patients who had antibodies in their serum, as well as recently transfused patients.10
The Microarray large-scale genotyping technique can be considered a rapid and high-throughput genotyping method to support the blood donor selection and the patient care quickly and efficiently, since the serological determination of multiple antigens of large groups of donors becomes expensive, laborious and hampered by the lack of antisera. However, this method is best applied at large hemotherapy centers, since this assay requires considerable sample accumulation, making it less attractive for solving individual cases for emergency transfusion care. Conventional PCR techniques are a good option for routine blood banks, where the number of patients and donors is smaller, since they are fast, more economical techniques that have good sensitivity and specificity for the detection of the main blood group antigens.
Analyzing the advantages and limitations of the molecular and serological methods tested in this study, we note that both are important and complementary, contributing positively to the determination of the patient's phenotype. The possibility of performing genotyping in conjunction with hemagglutination changes the range of possibilities in transfusion procedures, thus increasing the safety of transfused patients.
Further studies can evaluate other methodologies, as well as seek alternatives that reduce cost while maintaining the quality of results. Further investigation is also required to evaluate the total cost of each method, as well as the specificity and sensitivity for the detection of each allele.
ConclusionDetection of erythrocyte antigens by the PCR-SSP proved to be more economically feasible, compared to other serological and molecular methodologies. On the other hand, when the time to obtain the results was analyzed, the serological method required less time than the molecular methods. Nevertheless, for the selected samples, all methods proved to be effective and safe in the detection of erythrocyte antigens, since there was no discrepancy among the results obtained.
Conflicts of interestThe authors declare no conflicts of interest.
We are grateful to collaborators from the LIG-UEM, Maringa Regional Blood Center, Francisco Beltrão Hemonucleo and Molecular Biology Laboratory of Hematology and Hemotherapy Center of the UNICAMP.